Machine Learning for Protein Design & Yeast Make Medicines from Nightshade (Issue #6)
Also This Week: Changes to this newsletter, assembly of 35 DNA parts at once, and open-source software makes measurements simple.
Good morning.
This newsletter has changed. Research items will now be curated, non-exhaustive, and explained in a few sentences, rather than provided as a list. I will still cover peer-reviewed research, preprints, and reviews. News items, and a special bonus section, have been moved to the end of the newsletter. While I hope that this narrative format is more appealing to readers, I welcome your feedback via direct Twitter message or comment.
🧬Featured Research
Biosynthesis of Plant-Based Medicines in Baker’s Yeast (Open Access)
In a tour de force of metabolic engineering, Prashanth Srinivasan and Christina Smolke at Stanford have engineered baker’s yeast to produce tropane alkaloids. These compounds, which are normally extracted from nightshade to treat neuromuscular disorders, were biosynthesized from starting sugars and amino acids. Constructing the synthetic pathway required more than twenty distinct proteins—taken from yeast, bacteria, plants and animals—targeted to six sub-cellular compartments, to produce the medicines. The study was published in Nature and has been widely covered in the media, including as a Nature News & Views story and in a Stanford press release.
Assembling 35(!) Pieces of DNA in a Single Tube (Open Access)
In 2018, I read a stunning paper by a team of scientists from New England Biolabs and Ginkgo Bioworks, describing an improvement to Golden Gate DNA assembly. In that paper, they described the efficient, accurate assembly of 24 DNA fragments in a single tube. Now, many of those same authors have built upon their prior study, reporting the routine assembly of 35 DNA fragments in a single reaction. To optimize the system, the authors used DNA sequencing to explore possible combinations of restriction enzymes, and then “incorporated these findings into a suite of webtools” to design more efficient DNA assembly reactions. This study was published in PLoS One.
Minicells, with Miniature Genomes, Can Build Themselves (Open Access)
Can a cell be built from scratch? A lot of people think so, and they may not be far off. A new study in Nature Communications reports that liposomes—basically sacs of phospholipids that enclose water—can be packed and programmed with a genetic blueprint. Liposomes were packaged with a loop of DNA encoding seven different genes, together comprising a biosynthetic pathway to build phospholipids. By using “fluorescence-based probes”, the researchers were able to “directly visualize membrane incorporation of synthesized phospholipids at the single vesicle level”—in other words, miniature cells, with miniature genetic programs, can help build themselves.
Fine-Tuning Gene Expression in Plants (Open Access)
Promoters are short sequences of DNA upstream of a gene that play a major role in determining how much of a protein is produced. But the rules underpinning promoters, and a gene’s expression level, have proven difficult to unravel. This week, the Patron lab reported an experimental system to rigorously investigate how different traits in a promoter—its sequence and positioning of regulatory elements, for example—impact the expression level of a gene. With these findings, a suite of “minimal” promoters were developed that can be used to more precisely tune a gene’s output. The study was published in Nucleic Acids Research. Read the press release from the Earlham Institute.
Software Package Simplifies Inter-Lab Measurements (Open Access)
The International Genetically Engineered Machine competition, or iGEM, has brought together young synthetic biology students for nearly two decades. A cornerstone of that competition is the measurement lab, whereby students carefully measure engineered cells and report their results to study how those measurements differ across laboratories and devices. A new study, in ACS Synthetic Biology, builds upon the dire need for standardization in synthetic biology, offering a software tool (written for R) that can calibrate fluorescent and plate reader measurements.
🧫 Rapid-Fire Highlights
More research & reviews worth your time
The first eukaryotic MAGE method was published in Cell in 2017, but improvements are finally here. The new version of MAGE, which can be used to implement many edits to a genome simultaneously, improved “editing frequencies up to 90%, reduced workflow time by 40%” and lowered “the rate of spontaneous mutations ∼17-fold”. It’s basically genome editing on steroids (bioRxiv). Open Access.
A CRISPR-based “chromosome drive” in yeast has been used to remove a synthetic chromosome X via a single, double-strand break in the DNA. A wild-type chromosome X, harboring a fluorescent protein or an entire biosynthetic pathway, was then duplicated by sexual reproduction, demonstrating that chromosome drives can be used to selectively transmit genetic information in yeast (Nature Communications). Open Access.
A CRISPR-based transcriptional repressor has been implemented in mice to modulate the immune system. Repression of a gene called Myd88 “can act as a prophylactic measure against septicaemia in both Cas9 transgenic and C57BL/6J mice” (Nature Cell Biology).
By deregulating central metabolism in E. coli with CRISPRi and targeted proteolysis, the Lynch lab at Duke was able to more scale up the production of “alanine, citramalate and xylitol, from microtiter plates to pilot reactors”, with engineered bacteria (bioRxiv). Open Access.
A new review on the molecular mechanisms of CRISPR-Cas bacterial immunity was published by Nussenzweig & Maraffini (Annual Review of Genetics). Another review espoused the promise of anti-CRISPR proteins for building gene circuits (Frontiers in Bioengineering and Biotechnology).
The Church lab is using machine learning to guide protein engineering efforts. In a new preprint, they claim that “as few as 24 functionally assayed mutant sequences” can be used “to build an accurate virtual fitness landscape” (bioRxiv). Open Access.
DNA origami nanostructures are used to “view” objects with nanometer-scale resolution. The method can write distances into DNA molecules, and use sequencing to reveal the geometry of tiny, molecular geometries (bioRxiv). Open Access.
A study reported a new way to find baker’s yeast with high nucleic-acid content by monitoring rRNA synthesis. The method uses a simple fluorescent readout, and flow cytometry, to find cells with more nucleic acids (Microbial Biotechnology). Open Access.
An intriguing review on gene delivery for the skin was published. As companies like Azitra look to engineer the skin microbiome, viral delivery methods may not be the best option (Trends in Biotechnology). Open Access.
Caulobacter cells were engineered to secrete an extracellular matrix protein fused to SpyCatcher, enabling “programmable extracellular protein matrices” (bioRxiv). Open Access.
A new method was used to measure the growth of different strains within a microbial community, using a combination of genome editing and a whole-cell biosensor (Synthetic Biology). Open Access.
A mathematical model outlined the molecular rules that scientists could use to program cells to form specific shapes. The study predicts that a single molecular signal can coax cells into forming a rod shape, while multiple signals can be used to create more complex geometries (bioRxiv). Open Access.
A full issue on yeast synthetic biology was printed, featuring articles on mitochondrial engineering, converting starch to ethanol, and transposable elements in baker’s yeast (FEMS Yeast Research).
Scientists report the computational design and creation of biological walking machines. The biobots can move about and maneuver themselves (bioRxiv). Open Access.
If you want to release GMOs into the wild, you’ll want to consider how to stop them if they grow out of control. A new study swapped codons in E. coli to create a “genetic firewall” that prevents leakage of engineered DNA into the environment (ACS Synthetic Biology).
📰 #SynBio in the News
In the biggest news story this week, a global commission has laid out a roadmap for the genetic editing of human embryos. The news was covered in many outlets, including WIRED, MIT Technology Review, STAT, Nature and Science. Despite the risks outlined by the commission, a Russian scientist plans to proceed with his ‘CRISPR baby’ experiments.
In more CRISPR news, The Scientist wrote about how synthetic biologists are scaling up COVID-19 testing.
Ethicists at NYU expounded the dangers of DIY vaccines in an op-ed for Science magazine, taking aim at George Church and other scientists that were featured in Antonio Regalado’s MIT Technology Review article last month.
Roya Aghighi and scientists at the University of British Columbia have created Biogarmentry, a living textile made from algae, which was featured on CNN Style.
San Diego-based Algenesis is making biodegradable, algae-based polyurethane, as featured on Fast Company.
Spectrum covered a study (published Aug. 1) that used a Cas13-based system to degrade mRNAs, which could prove useful for autism research.
NIH awards $14.6 million for HIV gene therapy research to scientists at USC and the Fred Hutchinson Cancer Research Center.
Yeast were extracted from a loaf of bread buried around 2000BC and used to bake a modern loaf. It’s not synthetic biology, but I have a soft spot for baking!
🐦Science Threads on Twitter
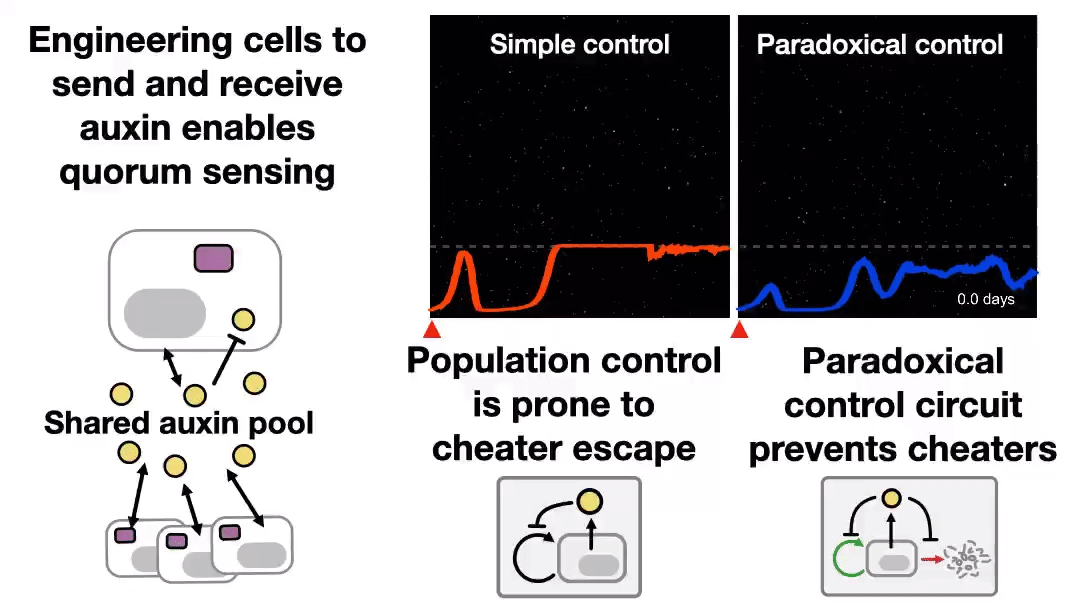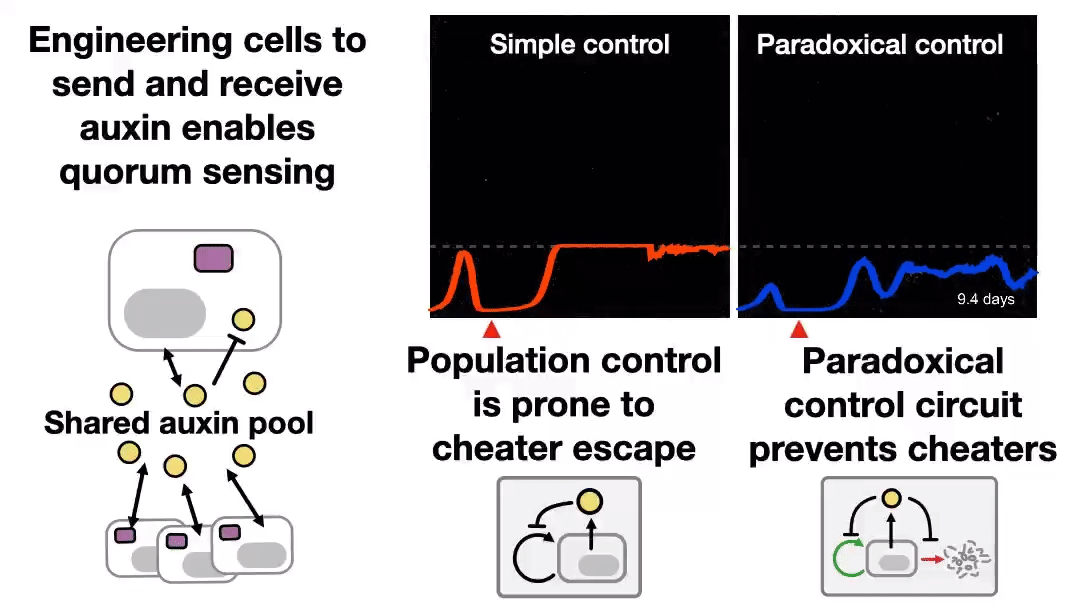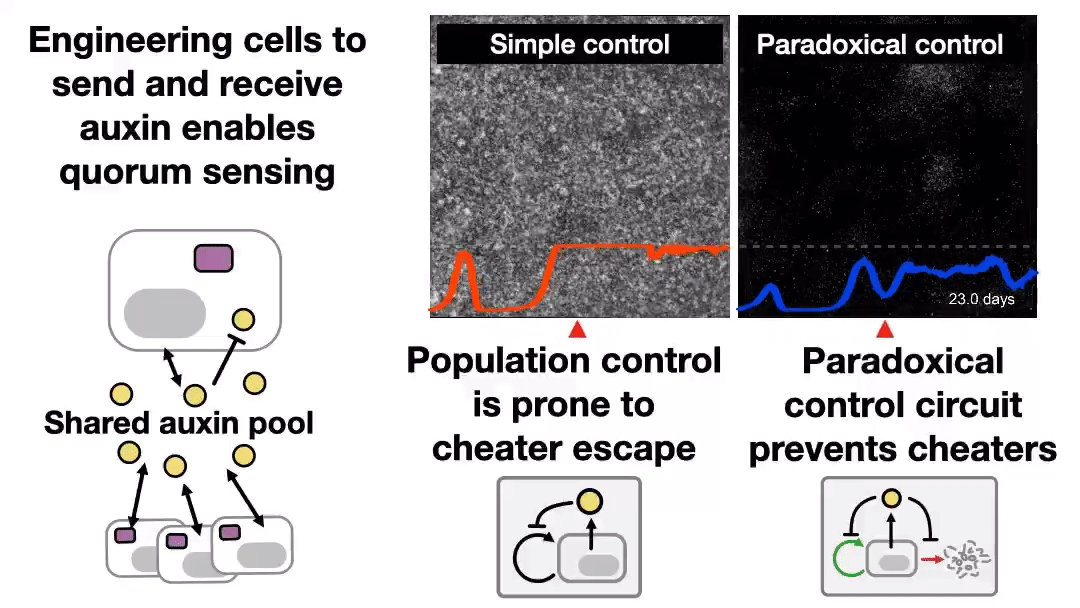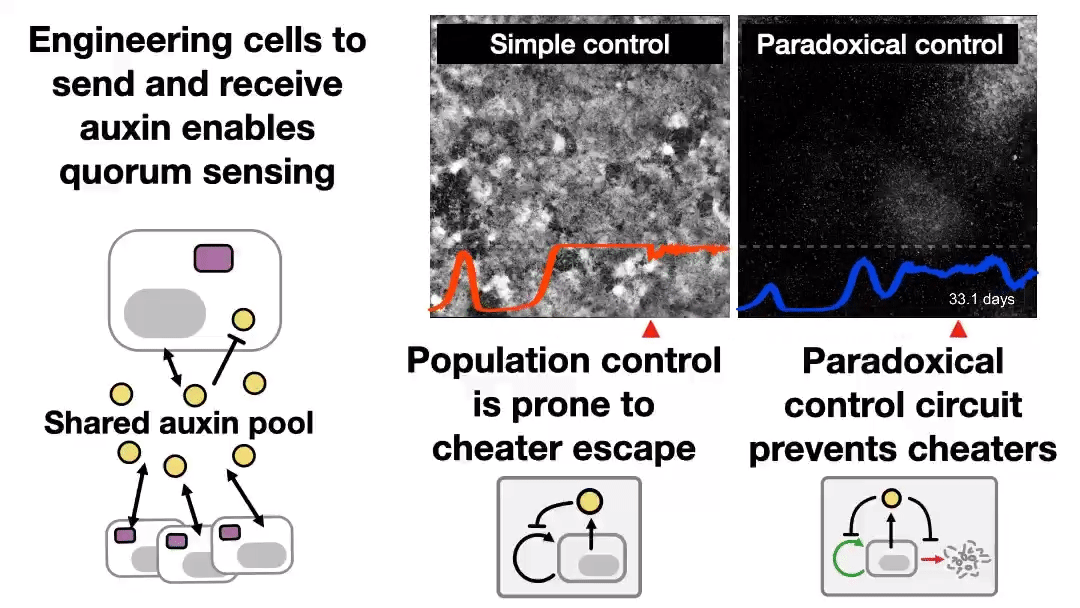
A new preprint was uploaded to bioRxiv last night, describing a synthetic system to control cell populations, without “breaking”, for 43 days! Read the Twitter thread explaining that study 👇
Thanks for reading This Week in Synthetic Biology, brought to you by Bioeconomy.XYZ.
A version of these newsletters is posted on bioeconomy.xyz and my website, hiniko.io. Reach me with tips and feedback via Twitter direct message at @NikoMcCarty.