I. Dreams
This essay is a thought experiment about a protein printer, a new technology to convert digital bits into physical molecules. It is speculative writing, because this printer doesn’t yet exist. But there are no physical reasons that it couldn’t, and we think it could change biology as much as printers changed the news.
We are living through an inflection point in biology’s history, in which a simple laptop can design proteins with wondrous new functions. Scientists at the Institute for Protein Design have crafted proteins that self-assemble into nanomaterials or that can be used as vaccines for the flu and HIV. In the future, “designer” proteins could be used to make new medicines or clean up toxic spills in the environment.
But “progress in science depends on new techniques, new discoveries and new ideas, probably in that order,” said Nobel Laureate, Sydney Brenner. And, unfortunately, it’s much simpler to design a protein than to build one. Computers are plentiful and electricity is cheap. Millions of proteins can be sculpted in the digital world, but each one costs at least $50 to make in the real world. That’s because proteins are made, in the laboratory, using synthetic DNA and cells; and DNA is expensive. Hundreds of protein designs must also be tested before we find one, finally, that works.
Our machine would make proteins without using any DNA or cells. It could be sold as a kit, and could theoretically make one billion unique proteins for about one dollar.
Much of the following text is speculative. We’ve tried to point out intellectual holes, but we hope you’ll reach out and leave comments. This isn’t a grant proposal; it’s just us speculating about how to solve a problem, much like we would at the bar on a Friday night. After all, that’s the place where many scientific ideas first take form and begin their slow journey toward reality.
Note from our sponsor
Fifty Years supports early-stage founders solving the world’s biggest problems. Their Fifty 50 program teaches entrepreneurial PhDs/postdocs in Bio or Climate everything they need to start a company. If you know someone who could be a fit, nominate them.
II. Polymers
A protein is a long rope, speckled with beads, and each bead is an amino acid with a distinct shape and charge. The order of beads determines the final form of a protein; sequence begets function.
Proteins act as machines and messengers inside crowded cells. Some catalyze reactions 30 million times per second. Others break down food and build up cell walls, or capture carbon and store it in sugar molecules.
All cells make proteins in two steps: DNA is transcribed into messenger RNA, which is then translated into protein. This is the Central Dogma, a concept taught in textbooks and whispered about by biologists in reverent undertones. It is the untouchable foundation of molecular biology. But we will try to convince you that, perhaps, we can do better.
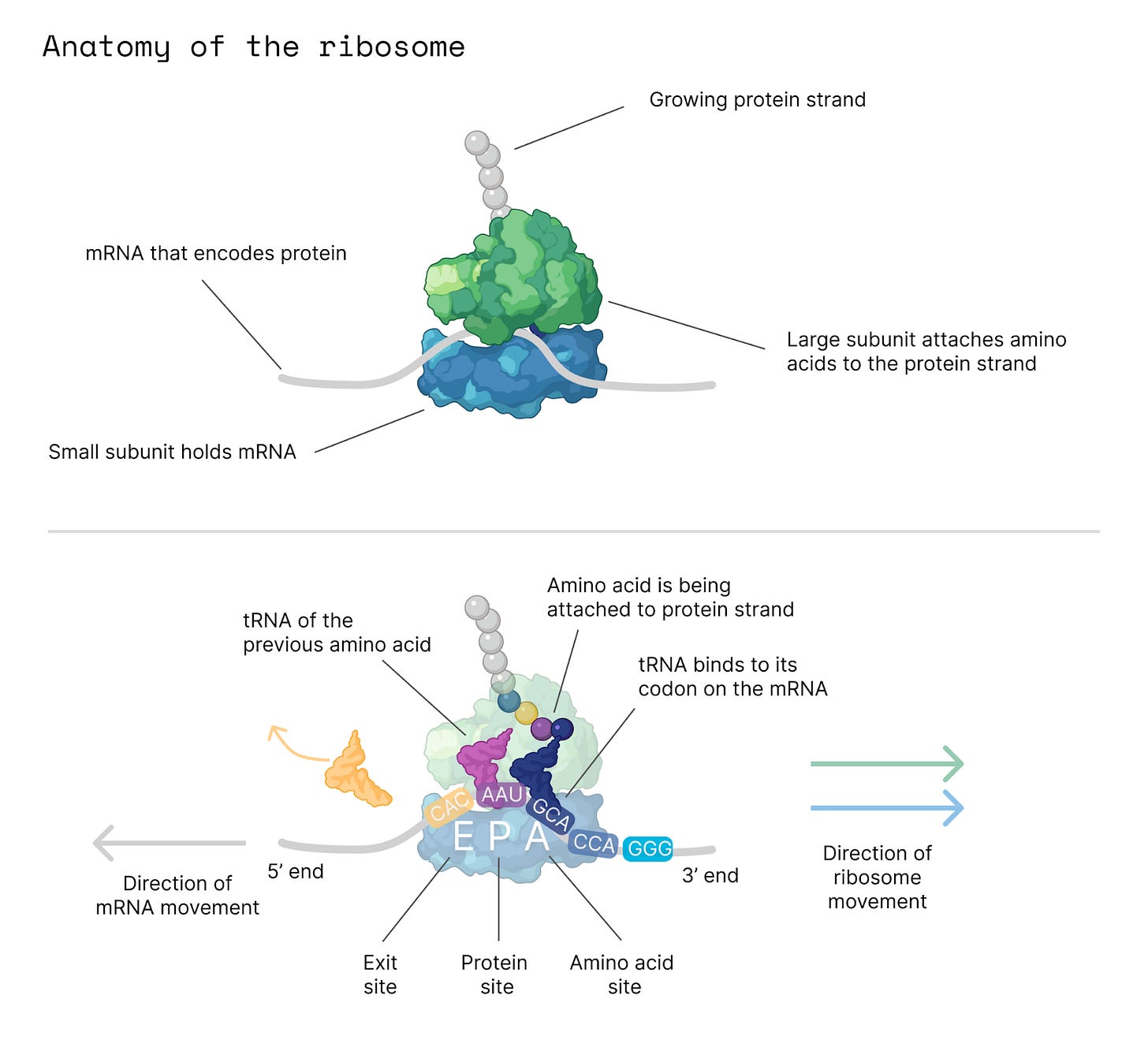
The ribosome is a molecular printer that threads ‘beads’ onto a protein string. It moves along RNA and reads three letters at a time, called codons, to build the amino acid sequence of a protein. The codon ‘GCA’ encodes alanine, ‘CUC’ encodes leucine, and so on.
Now, when we try to make proteins in the laboratory, there are only two options: Build with chemistry or harness the ribosome. The first option takes individual amino acids and fuses them together to form a chain. Chemists can make proteins with up to 164 amino acids in a few hours, but this requires specialized equipment and chemicals.
The second option, which is more common, is to design a protein on a computer, work backward to turn its amino acids into a DNA sequence, and then send that out to a chemical synthesis company. Scientists build the physical molecules and return them in the mail. The gene is then inserted into cells, and the ribosome does its thing and makes the protein.
But this is too many steps. It’s like writing before printers were invented. It’s like sending a text to a printing house, only to see your work weeks or months later. What we need, really, is a printer that takes words from a screen and quickly converts them into a physical book. In other words, we need to hijack the ribosome to turn bits into molecules, without DNA.
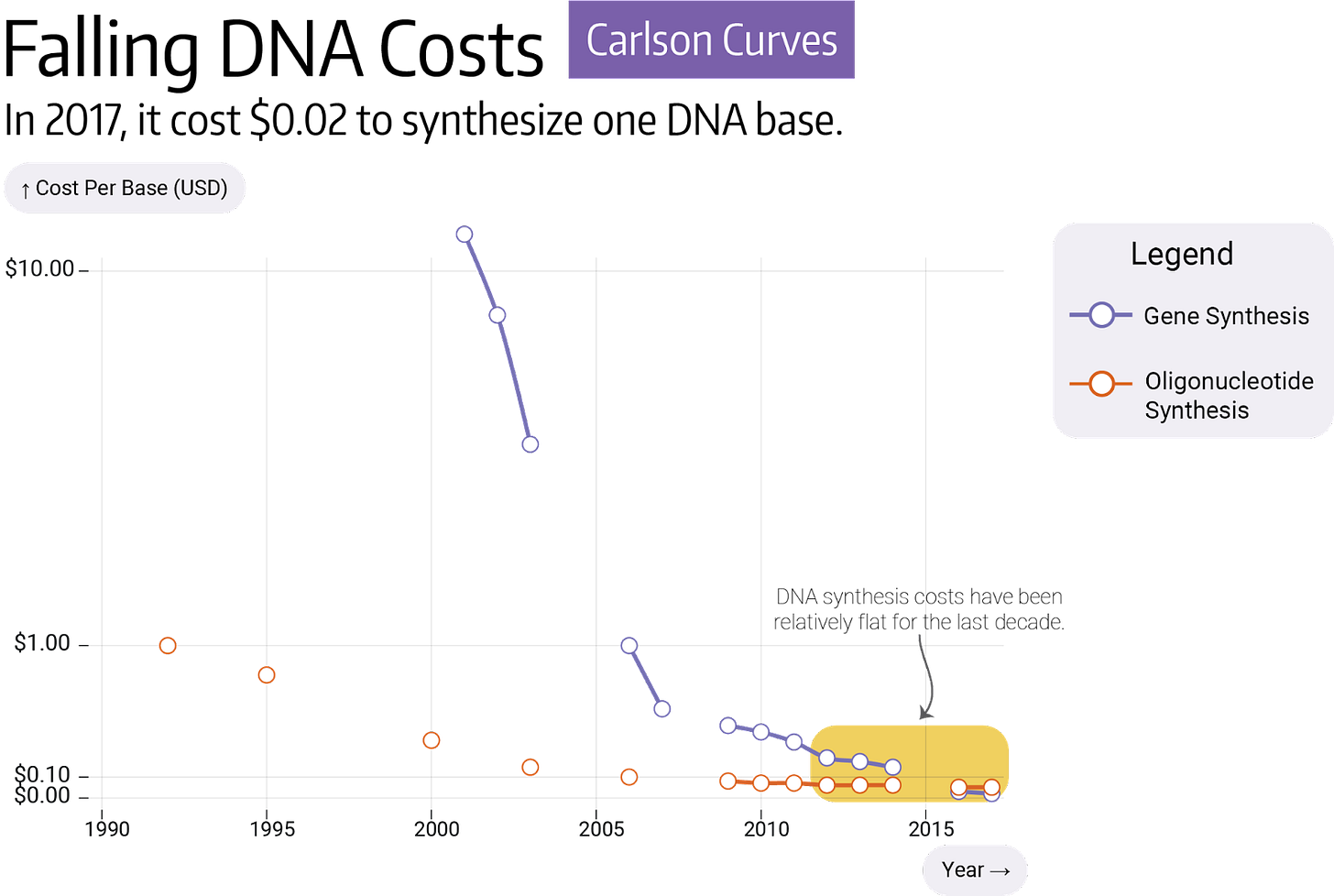
A typical protein has 400 amino acids, and each amino acid is encoded by three letters. This means that 1,200 bases of DNA must be ordered to build one average protein. Most DNA synthesis companies, such as Twist Biosciences, sell genes for as little as 7 cents per base, so a gene usually costs between $50 and $300. Many companies are building new technologies to make longer pieces of DNA, but we’re not aware of any that intend to cut costs.
Hence, our search for a cheaper and faster way to make proteins.
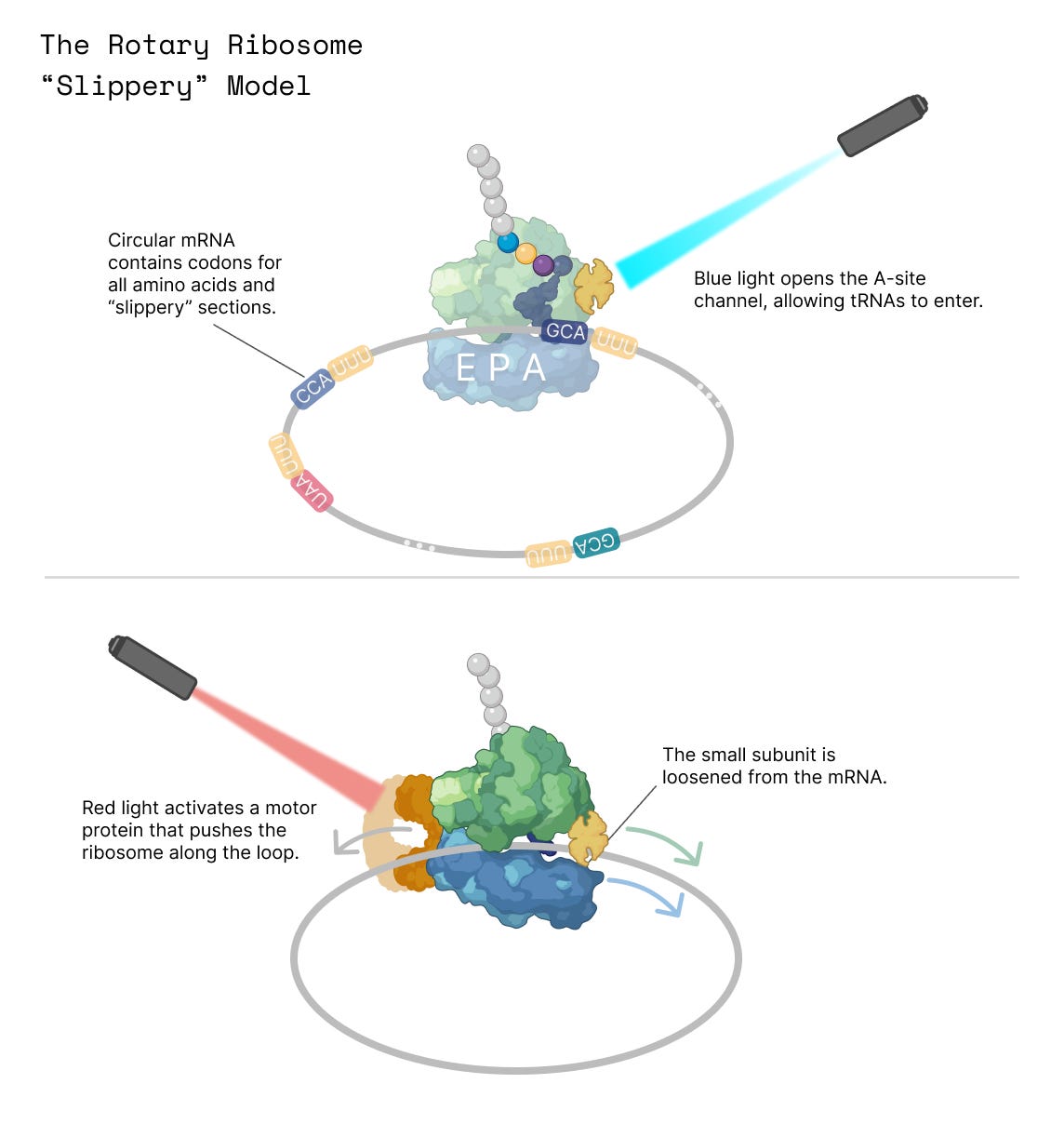
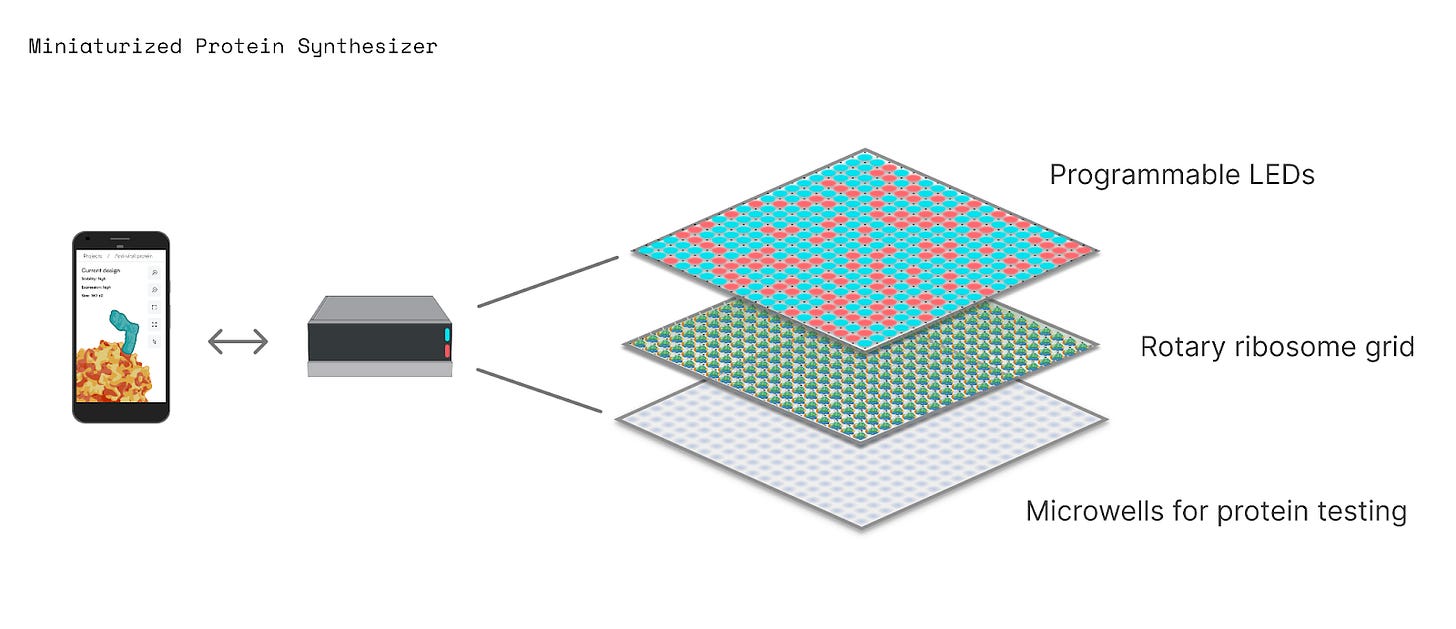
When we envision what a protein printer might look like, we think of a sort of rotary telephone in which all the different codons — AUA, AUG, AUC, and so on — are placed on a single, continuous loop of RNA. The ribosome would then be engineered to ‘hop’ from one codon to the next to build custom proteins.
This would require that we reimagine core parts of the Central Dogma, which won’t be easy. But we’ve workshopped the idea with ribosome engineers, and not a single person said, “This is impossible” or “This breaks a law of physics.” Two people sent emails with sketches of how this protein printer might work, and one said that his research group had thought about pursuing a similar idea.
So, here’s the plan…
III. Blueprints
The first step is to build a loop of RNA that encodes all the different codons. This is simple. We’ll start by making a linear RNA strand, using chemistry or biology, and then fuse the two ends together with a ligase enzyme. Circular RNAs are translated by the ribosome, just like normal RNA, but decay far more slowly because they are not easily recognized by “degrader” enzymes. Circular RNA has a half-life of 24 hours or longer, compared to perhaps a few minutes for linear RNA.
The circular loop does not have to contain all 64 possible codons, because the genetic code is redundant and there are only 20 amino acids. But the loop should have a diameter wide enough for the ribosome to freely move around.
Now, the next steps are more difficult to solve, because they require ribosome engineering.
Ribosomes are made from dozens of smaller proteins and RNA strands that come together in a brilliant symphony of molecular finesse. This machine evolved over billions of years to be incredibly precise, and to move along an RNA strand by one codon at a time. And we want to subvert this.
For simplicity, we'll focus on the bacterial ribosome, which is made of two subunits, called 50S (large) and 30S (small), that are held together with non-covalent bonds. Amino acids are carried into the ribosome on tRNAs, which enter the ribosome on one side and exit out the other. This happens about ten times every second. Fresh proteins emerge from the ribosome like beads threaded on a string.
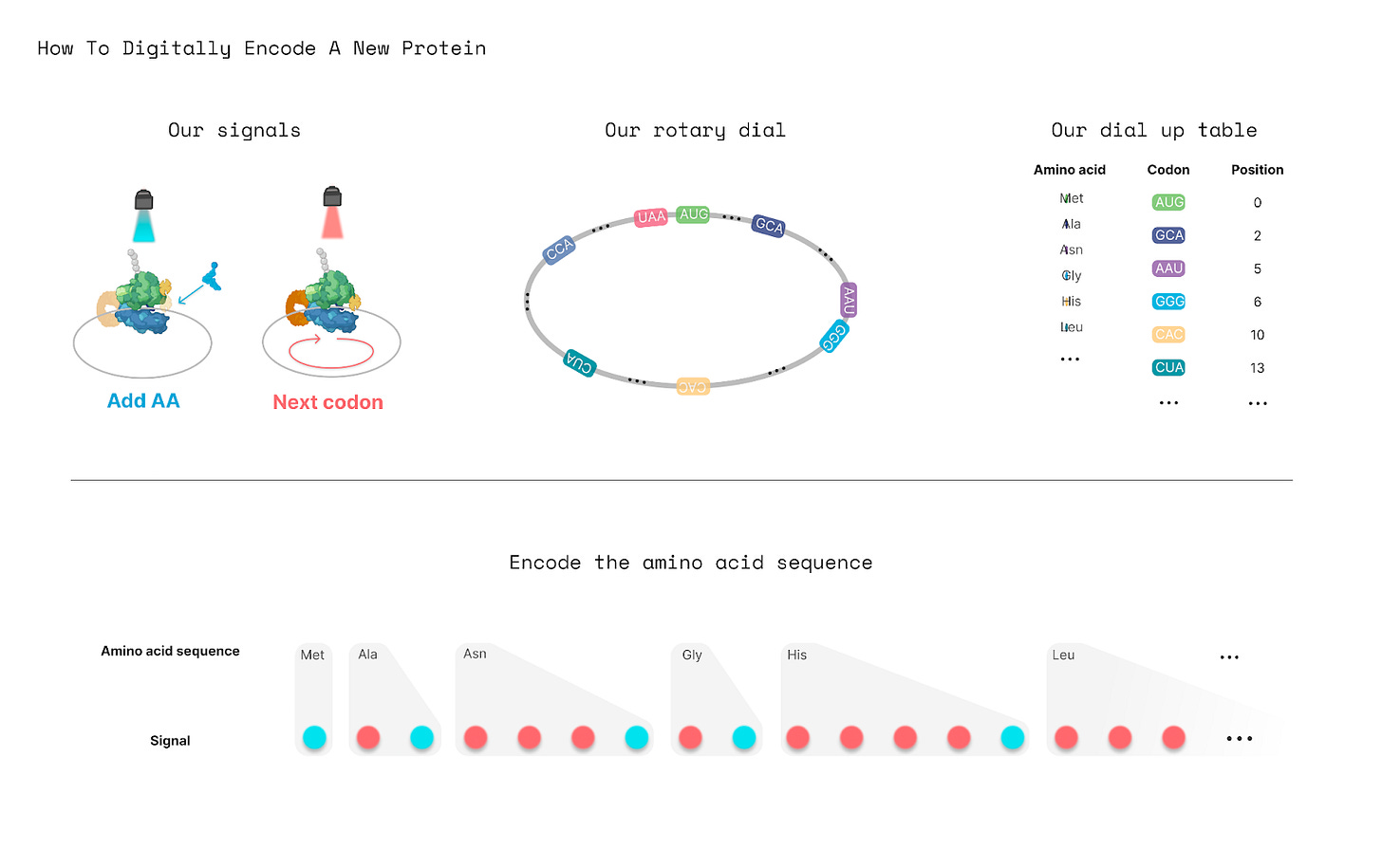
Now, for our machine to work, we have to carefully track the position of the ribosome as it moves along the loop. The ribosome should move to a specific codon, and then some kind of signal, perhaps a molecule or light, will signal, “Hey, add an amino acid at this position!” None of this will work if the ribosome falls off and we lose track of its position.
In a normal situation, inside cells, ribosomes grab onto short RNA sequences, called ribosome binding sites, to initiate translation. We’ll place one of these binding sites in the loop next to an ‘AUG’ start codon. The ribosome will grab onto this sequence, move to the start codon, and then will stay there, stuck, until we add tRNAs to its environment.
We’ll also tether the small and large subunits together — which has been done before — to keep the two ribosome halves together. A small protein “latch” would then clasp the ribosome together and keep it from falling off the RNA loop.
So now we have a ribosome that is stuck at the start codon on the RNA loop. How do we make sure that, as it moves along the circle, it only adds amino acids at the codons we specify?
This may be possible with a “protein gate” that physically blocks tRNAs from entering the large subunit. The protein gate could be clicked ‘on’ or ‘off’ using light. This sounds a bit like science fiction, but similar light-sensitive protein switches have been made in the past. It may be challenging, though, to make sure that only one amino acid is added each time.
Now, for the final part of our protein-making machine, we need to figure out how to make the ribosome “hop” or “slide” from one codon to the next. And this is no small feat.
“The ribosome is so engineered, so evolved, to not move in anything other than the three codon units,” says Jessica Willi, a biological engineer in Michael Jewett’s laboratory. “You may have to rebuild the entire small subunit to make this happen.”
*Gulp* Still, we think there are a few options to make this work.
Our first idea was to fuse the ribosome to a protein motor that burns chemicals to do physical work. Several DEAD-box proteins bind to RNA, and perhaps they could be fused to ribosomes to pull them along the RNA loop.
Another option is to hijack ribosome “sliding.” Sometimes, in nature, ribosomes get stuck on RNA or they have to skip certain codons. Excellent work from the Max Planck Institute shows that these ribosomes will stumble along an RNA strand — without adding amino acids to a protein! — until landing at a specific RNA sequence, where they resume translation.
Sliding starts when a “loop” in the RNA pushes the ribosome into a hyper-rotated state. A protein called EF-G then “fires” and burns energy to push the ribosome along. Perhaps we could hyper-rotate the ribosome artificially, using light (at a different wavelength than the protein gate) or another signal to coax the ribosome into this “gliding” state.
Each codon in the RNA loop would be placed next to a sequence that stops the ribosome from gliding, or surrounded by a hairpin structure that physically blocks the ribosome from moving further.
When the ribosome stops at each codon, a blue light signal would trigger it to add an amino acid, whereas a red light signal would push the ribosome onto the next codon without adding an amino acid.
None of these options are perfect, of course. We are fighting billions of years of evolution. The small subunit is exquisitely sensitive; it acts as a sort of brake pedal to stop the ribosome from slipping, says Alan Costello, a ribosome engineer, and so we’d have to loosen the small subunit’s tight grip on the RNA.
The tolerance for errors in this protein printer is also low. A typical ribosome makes a mistake once for every ten thousand amino acids. There has been a lot of recent progress in ribosome engineering, but we’re likely still years away from designing such an intricate, molecular machine with anything matching natural levels of precision.
But, fortunately, it doesn’t have to work perfectly straight away. Methods improve, slowly, over decades (just see the story of PCR, which was awful in the beginning). This protein printer could be tested, initially, by coaxing it to make small proteins with just a few amino acids. There is a tiny protein, with just 12 amino acids, that emits a fluorescent signal (manuscript in preparation). That means we could make many variants of the protein machine, put each of them into a different tube, program them to make this tiny protein, and see which tubes give off the most fluorescent signal.
IV. Plans
This essay is speculative. But by pondering theoretical solutions to a problem, we often think more deeply about the nature of the problem itself.
Remember that protein design has three steps: Design the protein, build it, and make sure it works. The first problem will likely be perfected in the near future, using some fancy AI models. This essay deals with the second problem, but it does not touch on the third. Protein design will only reach its full potential when the second and third problems get solved.
Adaptyv Bio, the company that Julian (a co-author of this essay) co-founded, is trying to solve the third part. They are building a foundry, where scientists can submit protein designs via an API and have them tested on automated workcells. By miniaturizing protein synthesis and characterization, and removing manual steps from the process, they aim to bring down the cost of protein engineering and help more people design proteins.
Still, the major obstacle in testing more proteins is the cost of DNA. So let’s see how our method compares.
A living cell burns 2 ATP and 2 GTP for each amino acid added to a protein. It costs about $1 for one milligram of GTP and about $1 per gram of ATP (a negligible cost). One gram of amino acids costs about one penny.
Let’s assume that our device requires 1,000 molecules of GTP and ATP to make an average protein. One could theoretically make 1 x 1018 proteins, then, for a thousand dollars. Even if you make a million copies of each protein, two trillion different kinds could be made for the same amount of money.
If the protein printer can be controlled with light, then its uses become even more profound. In the future, we may be able to design a protein on a computer, use a Python script to convert its amino acid sequence into a series of flashing lights, and then use those lights to move ribosomes to make proteins in real-time.
A student could even use LEDs, attached to an Arduino board, to control protein synthesis in a little tube. Or, a matrix of 64 x 64 LEDs, which costs about $50, could program protein synthesis in 4,096 wells at once. Light-based manufacturing is an established field, and more complex technologies are already used to make computer chips and 3D-printed parts.
But who is going to fund all of this? Maybe Speculative Technologies or Schmidt Futures have the money and desire. Or, perhaps, you are reading this and know how to make it all work — a solution has floated into your mind — and you’re buzzing to get started. We hope that’s true, because it would validate the entire notion that speculative writing can still drive progress forward.
So what are you waiting for? Let’s give it a shot.
Julian Englert is the CEO of Adaptyv Bio. Check out their blog to learn more about what the team is building to make protein engineering easier.
Cite as: McCarty, N. & Englert, J. A Protein-Making Machine. 2023 July. Available from https://www.readcodon.com/p/machine
Thanks to Erika DeBenedictis, Alan Costello, Michael Jewett, Devon Stork, Camila Kofman, and Jessica A. Willi.
Disclosure: The views expressed in this blog are entirely my own and do not represent the views of any company or university with which I am affiliated.



A few notes:
Light-based control of enzymes is a popular fount of...speculation, but too unreliable for the requisite level of precision. You might get a fast on-rate for, say, azobenzene switching, but the relaxation/off-rate will be too slow and stochastic. Do you have a method in mind for detecting when the ribosome has arrived at the desired site, or has traversed up to 19 "wrong" sites? A single protein molecule product is useless, so this needs to be synchronized between (at least) millions of ribosomes, some of which may proceed faster or slower over the template than others. This would also make detection of e.g. fluorescent excitation or quenching events unreliable as a method of determining ribosome location.
You mention the ribosome "gliding" around on the template, but not relying on the hypothetical motor protein for this movement, as the motor protein is used to melt through the blocking hairpin(s)? EF-G doesn't really act as a motor to continually translocate the ribosome forward; it only acts to push the A-site tRNA into the P-site after elongation. Since you're not continually adding amino acids as the ribosome glides, I don't think you can rely on EF-G to move the ribosome forward repeatedly. I wouldn't recommend using some kind of mutated EF-G for this purpose since it'd interfere with the actual elongation process. If we're going to be speculative, perhaps you could have some kind of nanotechnological "turbocharger" on the motor protein, so that it can provide both the forward gliding and the hairpin-overcoming.
The A-site "door" needs to be open long enough to ensure that the correct tRNA is added, and incorporated into the polypeptide, which varies heavily as the chance of the correct tRNA being inserted into the A site is somewhat randomly determined. It might be the first tRNA to enter the pocket, or the fiftieth. Leave it open too long, though, and you risk multiple incorporations, as you mentioned. Or, risk the ribosome sliding around on the template. You'd have to ensure that the ribosome is still able to perform some sliding, which allows the shuffling of the A-site tRNA into the P site during elongation, so you can't simply clamp it in place for a while.
I wouldn't say the the ribosome only moves codon-by-codon. Sliding usually ends at a site on the mRNA that matches the P-site tRNA's anticodon, which may include frameshifts. This P-site codon-anticodon interaction is fundamental to stabilizing the ribosome reaction, though I suppose not an insurmountable challenge. Similarly, after you've shoved the ribosome(s) to the correct site, the P-site tRNA is whichever was previously added to the protein, so unless there are 400 sites (20 x 20, one for each combination of P-site and A-site codons), the P-site tRNA won't match whatever codon it's sitting on.
Circular mRNAs are certainly useful, and seemingly essential for this approach, but I wouldn't highlight stability as a primary benefit. Since your reaction is running in vitro, you don't need to worry about RNA-degrading enzymes - unless you're having contamination issues.
It would seem that a better method of DNA synthesis, in terms of speed, cost, and length of contiguously synthesized DNA, would mostly solve the issues this proposal is trying to overcome. I agree that most new DNA synthesis companies aren't trying to compete on price, only length or turnaround time. Until we get past the crude hack of assembling hundreds of oligos together, that won't change, though we are working on that.
Sounds cool, thanks for sharing this!
I was wondering how you may re-set the device, to bring all ribosomes back to AUG before starting a new cycle. Perhaps you could use a third wavelength to trigger motion from the start codon (and maybe a fourth for the stop codon..?), so that when you’re done synthesising your protein, you can just slide everything forward e.g. 100 times with the standard wavelength for sliding; in this way all the ribosomes should slide until they reach the start codon, which they can’t leave without the special wavelength. Now your device should be ready to start over with a new protein :)
(Please let me know if it’s not clear!)