You’re reading Codon, a newsletter about the bio + tech advances enabling a brighter future for humanity. Subscribe to support my work.
In the 1500s, a Belgian alchemist named Jan Baptist van Helmont planted a willow sapling in 200 pounds of earth. After five years of dousing the plant in water, the tree grew 34 times in size, but the soil’s mass remained nearly the same. “Hence one hundred and sixty-four pounds of wood, bark, and roots have come up from water alone,” he wrote.
Van Helmont’s experiment is a small piece of the two-thousand-year history of alchemy. Newton spent nearly all of his later years searching for the elusive philosopher’s stone, believed to turn various metals into gold. And Paracelsus, the legendary Swiss physician, invited alchemists to teach his medical students and thought life relied upon sulfur, salt, and mercury, representing the body, spirit, and soul.
Alchemists aimed to transmute base metals, usually “sickly” iron and lead, into gold. Their pursuits sound foolish today (transmutations happen in supercolliders, which accumulate thin films of precious metals, like gold, over time), but many alchemists were celebrities in their own eras.
Fortunately, we no longer need to rely upon flimsy experiments to manipulate atoms. There’s a better way to rearrange and build molecules from raw materials. It’s called biology.
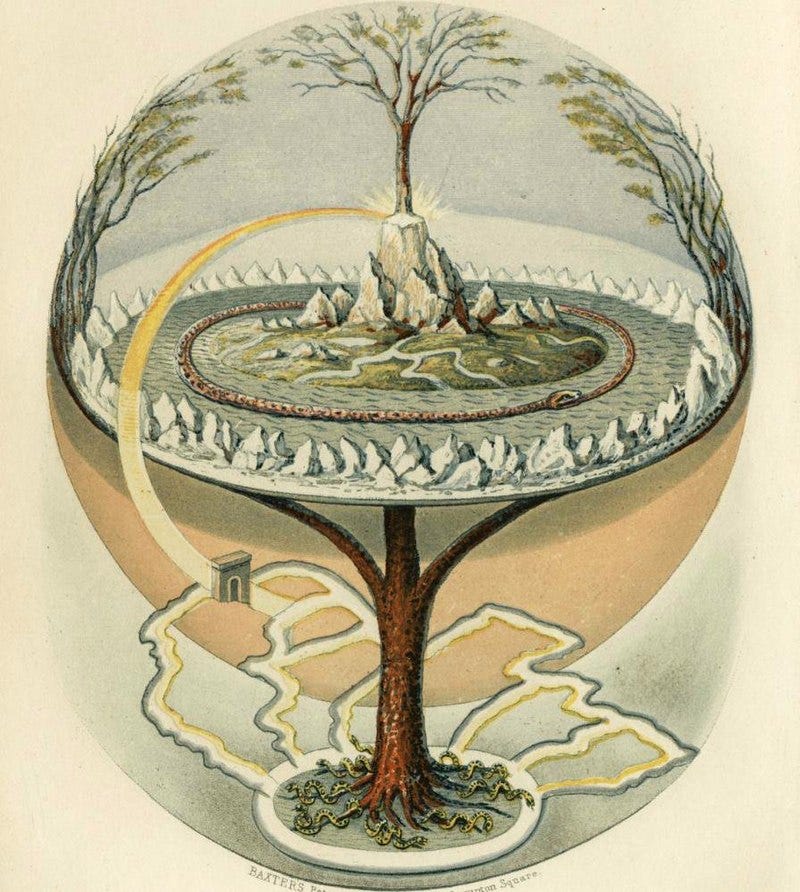
Every organism on Earth uses nanoscopic tweezers to cut apart and rearrange atoms. The most miraculous products arising from such jiggering are trees, which use little more than atmospheric carbon dioxide and water to build translucent leaves, sugars, and deep roots buried beneath soil. In other words, Atoms are Local; life harnesses its environment to grow, divide, and create. And biology does this at every scale, from individual proteins to entire continents.
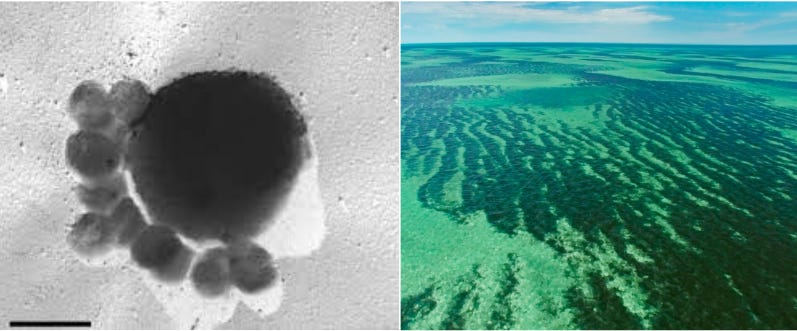
The smallest living thing is Nanoarchaeum equitans, an ocean-dwelling cell that was first spotted in a hydrothermal vent off Iceland’s coast. It measures 0.4 micrometers across, about 125 times smaller than the thickness of a sheet of paper. The world’s largest organism is a seaweed clone, called Poseidon’s ribbon weed, which is 4,500 years old and stretches across Australia’s Shark Bay.
Living creatures, out in the wild, take all forms and sizes. They can grow on one blade of grass or cover entire forests. But when we try to engineer biology at similar scales, our feeble designs often fail.
Just look at the story of synthetic silk. Multiple companies in the U.S., Germany, and Japan have been crafting synthetic spider silk for years, using yeast or other engineered cells to consume sugar and manufacture ultra-fine fibers. And yet, I still don’t have a silk jacket.
But this argument is misguided. It may not be easy to scale biology, but it is possible. It just takes a long time (decades) and most people are impatient. (When I lived in New York, cars would often honk at me immediately upon a stoplight turning green.)
Over the last fifty years — beginning with the rise of Genentech in the 1970s, and the cloning of the human insulin gene in bacteria — we’ve gotten much better at engineering and scaling biology. Entire synthetic pathways, made from proteins designed on a computer and created in a lab, capture carbon dioxide and convert the molecules into sugars in a test tube. AI tools have simplified protein engineering to an absurd degree. Newfangled chemistry is imbued into living cells as a matter of routine.
Biology at scale is no longer hype. Bioengineered products have reached commercial scales in every single scientific field, in ways that are often impossible with any other approach. Gene-edited viruses, first designed in a materials science lab, build ultra-efficient batteries. Engineered pigs, first created in an immunology lab, make humanized organs for transplants. Tweaked mosquitoes, pioneered in a zoology lab, curb diseases over hundreds of square miles.
In other words, biology is a foundational technology to build physical products from local atoms, and it is making a distributed impact across much of science.
Jump in to discover life at every scale.
From Molecules to Organs
A few decades ago, before CRISPR gene-editing, many chemists dreamed about ways to make proteins with new functions, found nowhere in nature. In 1984, a German chemist named Manfred Eigen proposed a plausible approach: What if enzymes could be evolved, synthetically, in a lab? DNA would be randomly mutated to create protein mutants, he imagined, and those mutants could then be ‘screened’ to find variants with a desired function.
But Eigen swiftly encountered an intellectual roadblock: it “would be hard if not impossible,” he said, to pluck out a specific mutant from a library with billions of similar-looking proteins.
It didn’t take long for Eigen’s idea to bear fruit. A decade later, Frances H. Arnold, a chemical engineer at the California Institute of Technology, did the first successful directed evolution experiment. Her team evolved subtilisin E, an enzyme that cuts proteins at serine residues, to remain active in an unnatural, polar environment.
The approach was basically Eigen’s: Mutate the gene encoding subtilisin, isolate enzymes from bacteria carrying mutated copies, test the enzymes, and ‘reward’ the winners. After just four rounds of selection, and ten amino acid changes, “an enzyme variant with 256-fold higher activity than the wild-type enzyme…was created.”
Arnold’s experiment quickly diffused into the chemicals industry where, in 1999, Novo Nordisk took a peroxidase enzyme from a fungus and evolved it, using the same approach, to work at much lower temperatures. This enzyme is now used in laundry detergents around the world and can eliminate about 1,600 pounds of carbon dioxide emissions, per household, per year. Before this enzyme, about 90 percent of all energy used in washing machines went to water heating. Energy consumption in clothes washers dropped by 75.3 percent between 1990 and 2010.
Arnold’s achievement exists at a marginal scale: A single enzyme. Let’s now move one layer up, to an individual cell. At this scale, no achievement is greater than CAR-T cells for cancer therapy.
In the 1990s, Carl H. June, an immunologist at the University of Pennsylvania, was quietly studying T-cells and their antigen receptors. He discovered, along with other scientists, that these immune cells detect foreign invaders via proteins on their cell surface, called antigen receptors. These proteins bind to antigens on a pathogen’s surface and then signal the immune system: “Red alert! We’ve been breached!”
But what if one could create new antigen receptors, he thought, that bind specifically to cancer cells within a patient’s body?
In 2009, his team engineered mouse T-cells with custom-made proteins, called CARs (chimeric antigen receptors), and showed that the engineered cells have antileukemic properties. (CARs were initially proposed in the late 1980s, but were not clinically tested as a cancer therapy until 2006, in patients with ovarian tumors.) The next year, two cancer patients were dosed with the therapy, which works like this: First, draw blood from each patient and isolate their T-cells. Next, add genes encoding CARs, designed to bind little spikes on the tumor cells, into the T-cell genomes. Finally, pipe everything back into the body. The engineered T-cells are ‘programmed’ to circulate through the bloodstream, hunting and killing tumors.
A partnership with Novartis, formed in 2012, pushed the therapy through late-stage clinical trials. And the final results were shockingly good: “55% of patients with relapsed or refractory (r/r) B-cell acute lymphoblastic leukemia (ALL) who were treated with CAR-T cell therapy Kymriah were still alive after more than five years,” according to company data. The five-year survival rate, before Kymriah, was about 10 percent.
Five years ago, the FDA approved Kymriah to treat acute lymphoblastic leukemia in young people. The two patients dosed in 2012 have had decades-long cancer remission.
Now we move another layer up, to entire organs.
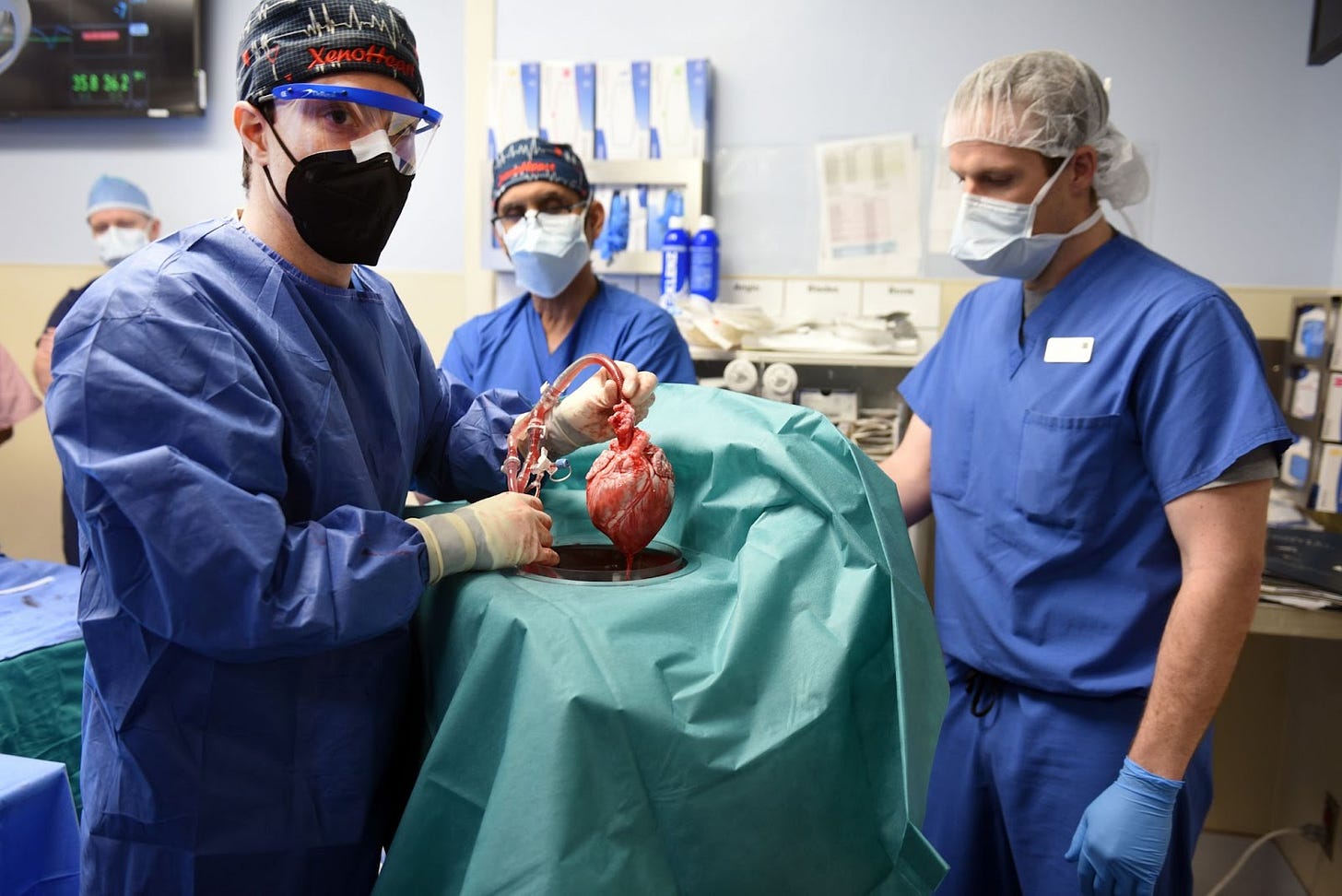
At any given time, about 3,500 people are waiting on a list to receive a heart transplant in the U.S. Pigs are an ideal vessel to grow human-compatible organs because the animals have short pregnancies, large litters, and their organs are roughly human-sized. But the scientific history of xenotransplants (xeno—, from the Greek xénos, or stranger) is fraught with failures.
In 1984, a one-month-old baby, named Stephanie Fae Beauclair, was born with hypoplastic left heart syndrome; the left half of her heart was severely underdeveloped, small, and feeble. A medical team in Loma Linda, California swooped in to save her, but couldn’t find a suitable human heart for the transplant. A surgeon, named Leonard Lee Bailey, opted instead to give Baby Fae a baboon heart. The procedure seemed successful at first, but Fae’s body rejected the heart and, 21 days later, she died.
Decades later, in the early 2000s, a small company named Revivicor developed gene-edited pigs, called GalSafe, which lack genes that cause hyperacute rejections in people. The pigs were developed together with surgeons at the University of Pittsburgh. (Revivicor, incidentally, is the same company that made the first cloned animal: Dolly the Sheep.)
The GalSafe pig organs are far safer for transplants than almost anything else that has been developed to date, but it took nearly two decades for the company to earn FDA approval. Last year, Revivicor published two clinical studies with their gene-edited pigs, transplanting kidneys and a heart into three separate people. The kidneys successfully made urine in brain-dead patients over a 54-hour period, without any side effects, while the heart went into a 57-year-old man who died a few months later.
That man, David Bennett Sr., did not die because the GalSafe organs are inherently unsafe. Rather, “Bennett’s heart was affected by porcine cytomegalovirus, a preventable infection that is linked to devastating effects on transplants,” according to reporting in MIT Technology Review.
Xenotransplants are booming on the back of Revivicor’s modest success. Companies in Cambridge, Massachusetts and Hangzhou, China are now making gene-edited pigs for organ transplants. Their technologies are similar — CRISPR and cloning — but each is spurring what will, undoubtedly, prove to be a booming organ industry.
Cocoons and Batteries
In 2020, a twisting white cylinder of snow-white silk draped the halls of New York’s Museum of Modern Art. Designed by Neri Oxman, a designer and professor at the MIT Media Lab, the artwork was crafted by 17,532 individual silkworms imported from Teolo, Italy. Over 10 days, these insects spun “a length of thread longer than the diameter of Planet Earth.”
It takes a lot of silk to make a tiny object. A single necktie is made from 250,000 feet of silk thread, and each silkworm makes about 2,000 feet of thread. Silk-making hasn’t changed much in the last few thousand years, either. It remains costly, technically challenging, and time-consuming.

A Swiss chemist, named Georges Audemars, was the first to earn a patent for ‘artificial silk,’ way back in 1855, “by dipping a needle into mulberry bark pulp and gummy rubber to make threads.” Modern approaches to make silk fibers with genetic engineering took off in the mid-1970s, when two Japanese biochemists successfully cloned silk fibroin genes into bacteria. To this day, synthetic silk remains the bien précieux for genetic engineers; a natural material, coveted by the wealthy, that can be made by microbes.
Even though silk is difficult to make, even in yeast, multiple companies are scaling quickly (the probability that I get a synthetic silk jacket in the next twenty years is not zero). AMSilk has grown production capabilities at a ~273% compounded annual growth rate since 2008. They currently make about eight tons of spider silk per year, but are expanding production to thousands of tons.
Silk is a low-hanging fruit because it is made with protein chains, and silk genes can be ported from silkworms into microbes. But there are other materials, far more difficult to create, that are found nowhere in nature.
Much like a chef’s recipe is consistent because it uses fixed amounts of standard ingredients, so too can biology fabricate nanomaterials, of a particular size and shape, by using DNA or proteins as physical rulers. A single twist of a DNA helix is 34 nanometers; biology relies upon exact standards.
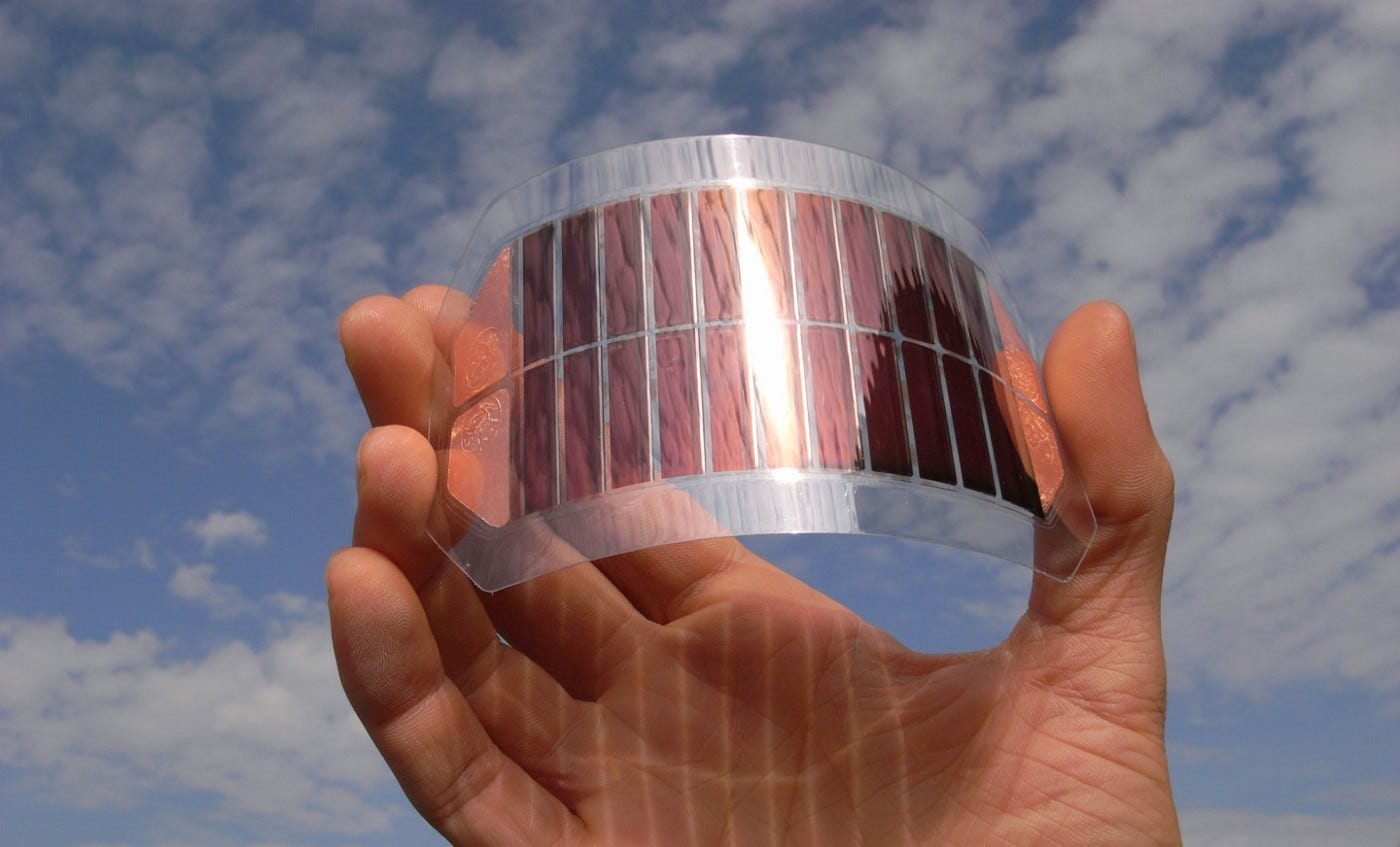
At MIT, Angela Belcher’s materials science group uses engineered viruses as physical rulers to build self-assembling materials for batteries, solar cells, and screen displays. A small change to a viral DNA sequence crafts materials with vastly different shapes or properties.
“Organisms learn to change what they use their proteins for. Instead of using them for certain functions inside the cells, they use them to grab elements from their environment and build exquisite nanostructured materials,” Belcher said in an interview. “Wouldn’t it be great if everything could be manufactured in that way?”
Belcher’s work relies upon a peculiar-looking virus, called M13, which resembles a long cylinder with small proteins dangling off each end. This specific virus is 880 nanometers long and 6 nanometers wide. By engineering the proteins on either end of M13 — which are each encoded by a separate gene — Belcher’s team has created self-connecting “LEGOs” that self-assemble into ordered sheets. These viral materials are used in lithium-ion electrodes and solar cells, and the viral proteins can also be modified to ‘grab’ onto gold or other metals to endow the materials with specific properties.
This technology — viral-mediated nanofabrication — has appeared in more than 50 patents. It’s the foundation of Cambrios, a startup that made touchscreen displays for Lenovo and Toshiba.
Biology is programmable and scales from single enzymes to entire organs and batteries. Now let’s go bigger.
Factories to Farms
The steel industry is dirty. Every ton of steel made releases about 1.8 tons of carbon dioxide into the atmosphere. But one man’s trash is another cell’s treasure.
Clostridium bacteria are found in the intestines of cows and people. These microbes feast on ruminant gas, including carbon dioxide and methane, and convert the carbon and hydrogen atoms into sugars and food. In some ways, cow gases, oddly, resemble steel factory waste.
In 2019, environmental scientists at the Oak Ridge National Laboratory and a company, called LanzaTech, engineered Clostridium bacteria to feed directly on factory waste gas and produce acetone as a byproduct. The key advance was simple: They added just two enzymes to the strains, which convert acetyl-CoA metabolites into downstream products.
The engineered cells achieved pilot-scale, cost-effective acetone production in 80-L bioreactors. Last February, LanzaTech and scientists at Northwestern University further optimized the engineered cells to manufacture both acetone and isopropanol in an entirely carbon-negative process. This is massively important for future climate biotechnology.
LanzaTech has partnered with steel factories across China to roll out its Clostridium strains. One of those factories made 9 million gallons of ethanol directly from steel waste gas in 2019, and the company closed a $500M funding round last year to scale even further.
Now we move up another level, from a single factory to entire cities or continents; this is where biology’s effects are most acutely felt.
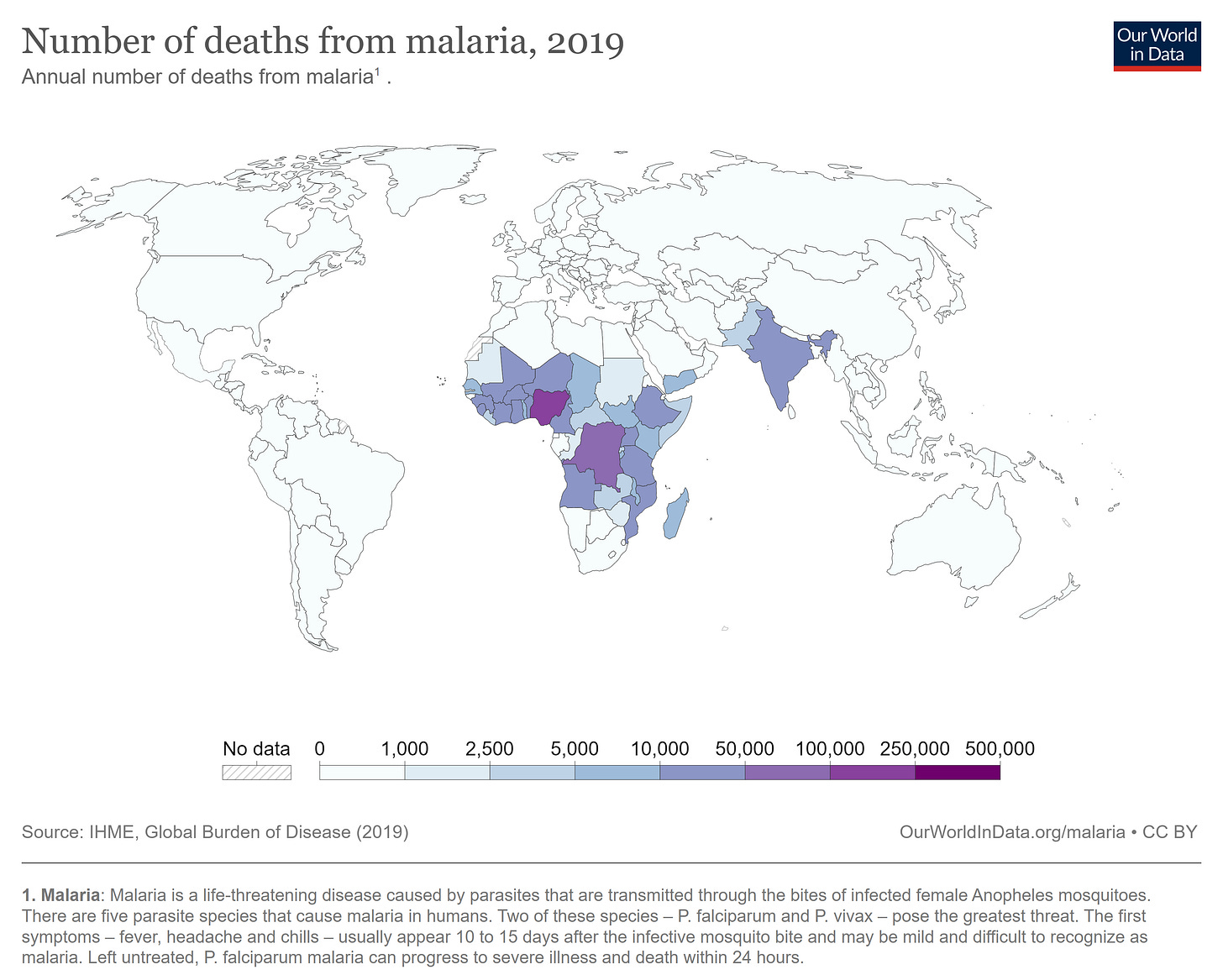
Mosquitoes kill millions of people every year. More than 50 million people contract Dengue fever each year, and another 600,000 die from malaria. Both diseases are preventable, and both hit the poorest nations on earth hardest. “Malaria has killed half of all the people who ever lived,” according to an uncited claim by John Whitfield. Even if that claim is false, mosquitoes are the deadliest animal of all time.
Here, too, genetic engineering offers a solution.
In the late ‘90s, zoologists at the University of Oxford developed a genetic system, in fruit flies, that kills off female offspring (only females bite people). The flies were engineered with a dominant lethal gene that is always passed on to children. This lethal gene also has an ‘on’ and ‘off’ switch; when the insects are fed a specific molecule (usually tetracycline), the deadly gene is shut off. But when these insects are released into the wild, in the absence of tetracycline, their progeny swiftly die.
The approach — which quickly made its way into mosquitoes — was a massive improvement over the classic sterile insect technique. In 1935, screwworm insects killed nearly 200,000 livestock in just a few counties in Texas. After World War II, some smart entomologists at the USDA, including Edward F. Knipling, bombarded male screwworms with x-rays, rendered them sterile, and airdropped billions onto farms across Texas. The sterile males had sex with females, which then produced defective offspring. Screwworms were eradicated by 1991.
The Oxford technology is more sophisticated than Knipling’s, and a British company called Oxitec has used it to great effect. Oxitec has released engineered mosquitoes into the Florida Keys, Brazil, and, soon, Djibouti. But progress is usually accompanied by criticism.
“With all the urgent crises facing our nation and the State of Florida…the administration has used tax dollars and government resources for a Jurassic Park experiment,” one critic told the Daily Mail. When I visited the Florida Keys to go deep sea fishing last summer, massive billboards along the main road echoed this sentiment.
Still, Oxitec’s insects really work. In May 2018, the company released engineered Aedes aegypti mosquitoes in four towns across Brazil, which suppressed the local mosquito populations by 89 percent or more compared to untreated areas. Oxitec also collected thousands of mosquito eggs from the Florida Keys to allay concerns that insects carrying the added genes will propagate indefinitely. Their studies “reported that all females that inherited the lethal gene died before reaching adulthood.”
Factories and mosquitoes are great, but biology’s crowning achievement — the irrefutable evidence that engineered organisms can address global crises — is food.
Rice is the staple crop for more than half of all people, and so it’s here that gene editing wields its greatest potential. Genetically modified rice has lifted millions of farmers out of poverty and staves off hunger for millions more.
Many of the world’s poorest farmers survive on less than $3 per day. About 25 percent of rice croplands flood at least once per year. Climate change is making this problem worse, especially in low-lying deltas across Vietnam, Myanmar, and Bangladesh. Flooding causes annual crop losses of more than $1 billion each year in southeast Asia.
Rice plants die if they are submerged in water for more than four days. A single flash flood, in these low-lying deltas, can bankrupt and starve entire communities. In the 1990s, a plant geneticist at UC Davis, named David James Mackill, mapped the ninth chromosome of a common rice strain and discovered the specific region responsible for longer tolerance of total submersion.
By the mid-2000s, Mackill and Pamela C. Ronald, another UC Davis scientist, discovered that the Sub1A gene was responsible for flood tolerance in rice. Engineered rice crops that carry this added gene could withstand total submergence in water for two weeks. By 2017, the modified Sub1 crops were grown by at least 6 million farmers in Nepal, Bangladesh, and India. They’re a lifesaver in flood-prone regions, those hit hardest by climate change.
This story always stirs deep emotions; I can feel it in my chest. How strange that one gene, Sub1, accounts for less than 0.1 percent of a rice plant’s genome, and yet tangibly improves the life of so many people. A single gene can help feed the world.

Ode to Patience
Biology scales and lifts millions of people out of poverty. It is a force for good.
The greatest barriers to biology at scale are no longer rooted in technological limitations because it has never been easier to design or engineer organisms. A few decades ago, synthesizing a single gene cost tens of thousands of dollars. It took H.G. Khorana and two dozen other scientists nine years, or a total of 200 person-years, to synthesize just 207 bases of DNA in the 1970s.
Today, we’re fast approaching a convergence point; a high school student with about 6 months of training can design DNA on a computer, model its behavior, clone the DNA, insert it into a living cell, and measure the cell’s output.
With fresh talent and bold ideas, massive problems in climate, food, and medicine — which once seemed impossible to solve — will swiftly fall. This is not hyperbole.
The next great challenges are unpredictable intangibles. Companies cannot scale because regulations are stifling, consumers want to see clear benefits from engineered products, and politicians don’t understand the technology. These issues will take copious amounts of time, and patience, to solve. Few scientists appreciate just how hard it is to make and market a consumer-facing product, deal with political turmoil, spend money to win over approvals, or jump through archaic federal regulations.
It took Revivicor nearly two decades to earn FDA approval. Breeders at the International Rice Research Institute have been making rice crops with improved flood resistance for nearly half a century.
Still, life works at every scale, from a single cell to seaweed that blanket ocean bays. It’s possible to harness biology’s potential, but the road is mostly unpaved. We need ambition, time, and patience.
Thanks for reading Codon. If you enjoyed this post, consider sharing it or subscribing. —Niko
Correction: An initial version of this article incorrectly stated that the world’s largest organism is a fungus in Oregon.




Awesome Substack! An ode to biology :)
Great read, glad I found this!