Genetic circuits control microbial communities
Yeast communities built using modular components behave according to simple, and predictable, mathematical equations.
Researchers have engineered a collection of yeast cells — each specialized for a unique function, like sensing small molecules or producing them — to communicate and work together in groups that execute complex tasks.
The new study, published in eLife, places particular importance on designing engineered cells with modular components, a property that allows genetic parts to be mixed-and-matched in different combinations to achieve distinct goals.
Modularity is highly-valued in engineering because modular parts do not change their properties when combined with other parts. Engineers build planes, trains, and bridges using the same types of screws and metal sheets. Such modularity helps engineers predict how well an airplane will perform in the sky, long before passengers shuffle into its fuselage.
Living cells are often programmed with synthetic genetic circuits; a set of instructions written in DNA that prime a cell to execute a particular program. Typically, a single cell carries all the components of a genetic circuit. That cell divides in its flask and, soon, there are millions of identical cells, all programmed to carry out the same function.
This ‘army of clones’ is a far cry from what happens in nature, though, where microbial communities commonly work together and rely on a division-of-labor strategy, whereby different cells perform specialized functions.
By mimicking nature, and breaking up genetic circuits into subsets of components placed into different cell types, bioengineers can build more complex circuits while reducing the burden borne by each cell. If parts of a circuit are at odds with each other, compartmentalizing them into separate cells can also prevent undesired interactions. Furthermore, this community-based approach can exploit synergies between its members, since some species in the group might naturally be better at certain tasks than others.
This new study fuses all of these concepts. It isn’t the first to engineer microbial communities, but the remarkable modularity of the engineered cells in this paper, specifically, makes it possible to predict, ahead of time, the behavior of varied cell combinations. Communities of yeast cells were designed on a computer and then built in the laboratory to perform complex functions; biological switches, logical operations, or living band-pass filters.
Bits and pieces
For the new study, researchers at the University of Washington and Virginia Tech first programmed yeast cells to communicate with each other by exchanging two chemical signals: the yeast pheromone, α-factor, and the plant hormone, auxin.
To do this, they constructed three different DNA modules and inserted them into the yeast. The first module senses a chemical signal. A second module processes that signal and determines whether the recipient cell should activate or repress its output. A third module determines the nature of the output: Either secrete a second signal, produce a protein to break down the current signal, or emit a fluorescent signal.
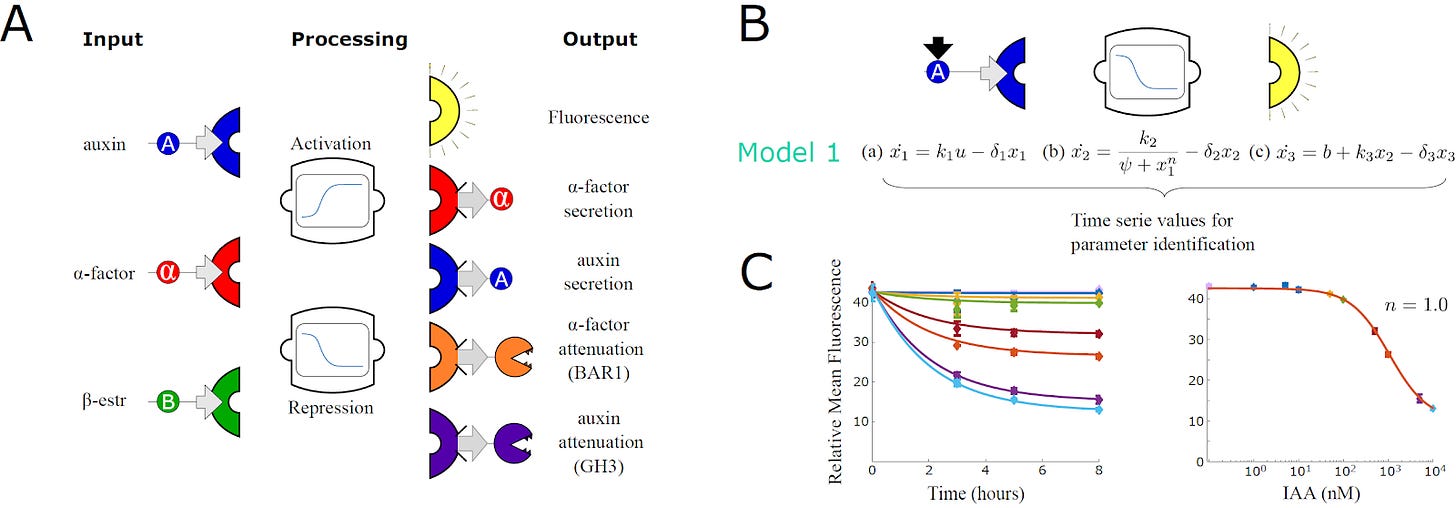
With these three modules, engineered cells interact with other members of the community and tune their own responses accordingly. All possible combinations of these three modules were constructed in the new study, resulting in a dictionary of 24 different yeast strains.
Start small
As a proof-of-concept, the researchers assembled all possible sender-receiver combinations. For example, sender cells that detect the small molecule, α-factor, and produce auxin in response were mixed with receiver cells that sense auxin molecules and emit fluorescence. The result is a simple, two-cell consortium that fluoresces in response to α-factor.
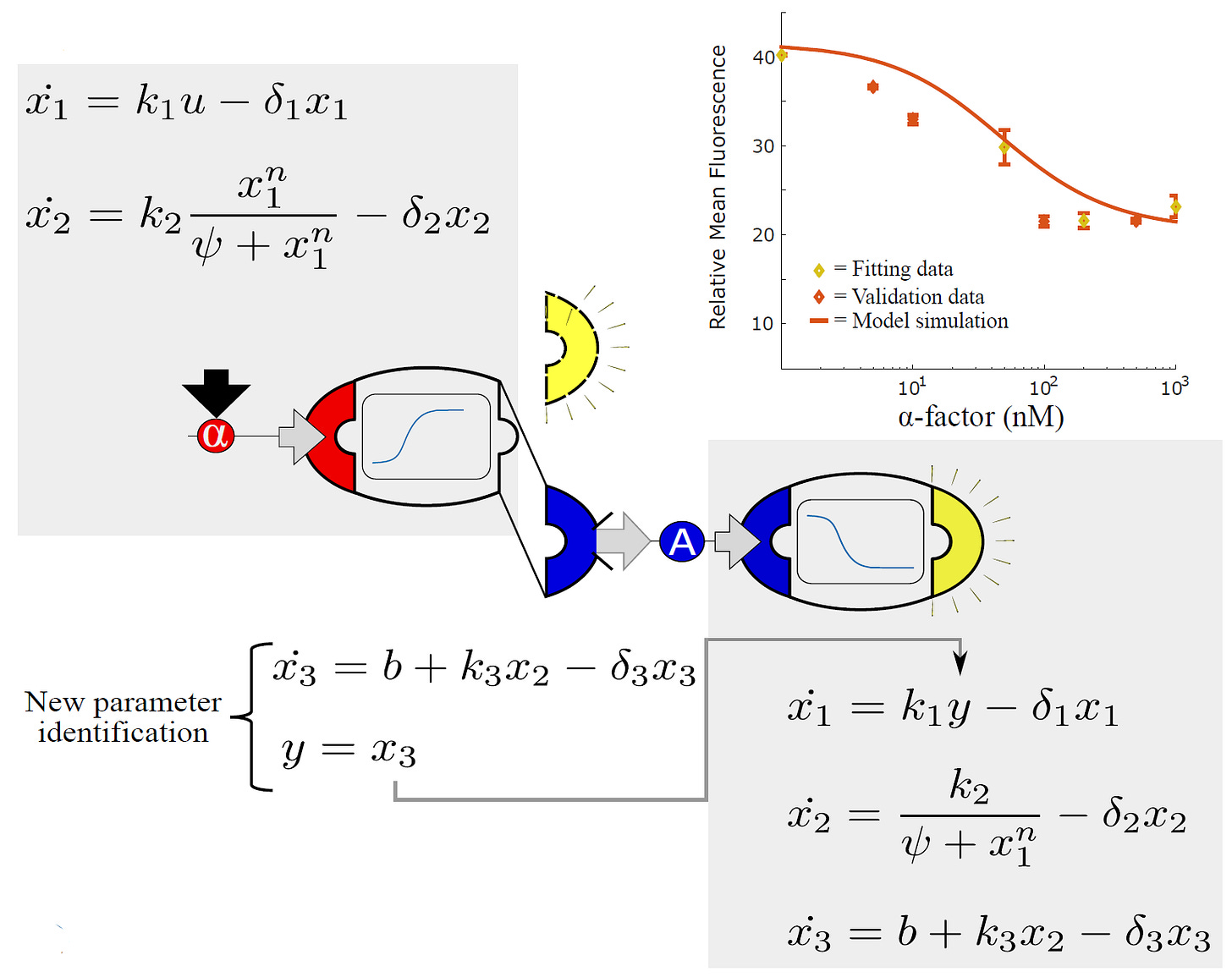
Since the cells are, themselves, engineered with modular DNA parts, so too are the mathematical models that describe their behaviors modular. Just three mathematical equations can describe a cell’s sensing, processing, and output modules.
Just as a sender-receiver community is assembled by combining two cell types, the researchers could also join the mathematical equations describing each cell to obtain a working model of the entire community. The output equation of the sender cells serves as input to the sensing equation of the receiver cell.
Quick switch
The sender-receiver communities had an almost linear response — a change in the input caused a proportional change in the output. For some applications, this is an asset, since it makes behaviors easier to predict. Some complex behaviors require dramatic, non-linear responses, though. An example of this is a bistable system, which can exist in one of two states (ON or OFF) at any given time. A switch can be flipped to revert the system from one state to the other.
To assemble a bistable consortium of yeast cells, the researchers first devised a strategy to generate non-linear responses in receiver cells that sense a signal. They constructed two new cell types: One produces the same molecule that it senses, while the other halts the degradation of a signal in response to the signal itself. As a result, these cells, in effect, amplify their input signals in a positive feedback loop.
A four-strain, bistable consortium was built with these positive-feedback cells. The resulting community produced a large amount of α-factor or auxin in a positive feedback loop and shifted between the two states when a quick ‘pulse’ of the opposite signal was added to their environment. Using mathematical simulations, the researchers also determined how much of each cell type should be added to the mixture to obtain an optimally-performing consortium.
This is another interesting property that arises when genetic circuits are distributed across cells; namely, the number of cells of a particular type determines the strength of its effect on the whole. In a conventional circuit, with only one cell type, researchers often ‘tune’ a circuit’s strength by adjusting the copy number of a plasmid harboring the genetic circuit’s parts. The physical amount of each yeast cell in a flask, though, is much easier to tune experimentally.
Pick-a-part
In a final batch of experiments, the researchers developed a computational pipeline that automatically designs the right community of cells for any given task.
The procedure simulates the behavior of all possible combinations of up to four cell types. The output of each simulation is assessed according to how well it accomplishes the desired task. This model dictates the relative amount of each community member to add to the flask.
Following this procedure, yeast communities that compare two inputs and produce an output according to predefined rules — so-called logic gates — were quickly designed and built. In another experiment, consortia that produce pulses of a chemical signal or that react only to intermediate concentrations of an input signal (also known as a band-pass filter) were quickly constructed.
Computer simulations, in this paper, revealed that less than 0.5 percent of all possible cell combinations could implement a band-pass filter. Attempts to identify these designs through trial-and-error would almost certainly fail. Moreover, exhaustive simulations can reveal interesting designs that would have been hard to imagine based on intuition alone.
Pitfalls
This study nicely demonstrates the advantages of modular, genetic circuits distributed over various members of a microbial community. However, there are still issues to be addressed before this approach can be used more broadly.
Only two independent communication channels were used in the study. Expanding the overall number of communication channels available would allow for greater complexity of genetic circuits.
And though an experimenter can set the relative amounts of each cell when assembling the consortium, each cell type’s proportion in the population will naturally change over time, according to how fast that cell type divides. Varied growth rates can probably explain why the ‘bistable’ consortium, in this study, lost its bistability after about 25 hours. Methods for stabilizing the composition of the consortia will improve performance over longer time periods.
Finally, the automated design pipeline simulates all possible cell-type mixtures to conceive the optimal design. Applying this approach to larger consortia will require clever computational methods to sort through an exponentially expanding number of combinations.
In spite of these limitations, this paper highlights the potential of synthetic biology for applying a strategy that is ubiquitous in nature and makes microbial communities successful: cooperation.
About the Author
A physicist turned synthetic biologist, Joaquín Gutiérrez is a PhD student at ETH Zurich, developing methods to control bacterial communities. His interests include genetic circuits, microbial ecology, evolution and discovering new interests. In his free time, he writes poetry and uses a piano to study his neighbor's stress response.
Reach him at joaquin.gutierrez@bsse.ethz.ch or on Twitter @GutierrezJoa

