
You’re reading Codon, a newsletter about the bio + tech advances that are building a brighter future for humanity. Subscribe and share.
Metaphors can misguide. When someone says that they “program living cells much like computers,” of course they’re leveraging simplicity to bring others up to speed on what’s possible with biology. And cells have certainly been programmed to do some crazy things — make concrete! capture carbon! detect landmines! But metaphors can also strip away a boatload of biological complexity. Cells are not computers. They’re way messier, infinitely weirder, and much more exciting.
Just imagine, for a moment, that you are a protein inside a cell. Perhaps your job — the one thing you’re supposed to do — is that you get spit out by a ribosome and then shove your face up into the cell membrane. You sit there on the outer edge of a cell, waiting for a chemical to breeze by. And when that chemical comes near, you grab onto it, shift and expand your protein body, and open up a pore into the cell.
Now imagine what it takes to engineer a new protein that does exactly the same thing, but opens up twice as wide. If biology were a computer, the fix would be simple: Add “2 *” to the code. But the reality isn’t simple. It’d probably take months or years of tinkering to get the new protein working in juuuust the right way.
It’s hard to “program biology” because cells are packed tighter than a family reunion at your grandmother’s single-story home. Just look at this painting from David Goodsell, a professor of computational biology at Scripps Research Institute, whose artwork captures the molecular denseness of a living cell. Every shape and squiggle is a protein, strand of RNA, fat molecule, or piece of DNA. That’s a lot of stuff.

It takes one protein in one human cell about 10 seconds to move from one side to the other. A single cell has about 2 billion proteins floating around inside. A single protein, moving through that cell, will crash and bump into other proteins thousands of times every minute.
All this to say: Metaphors are great tools to scale explanations. They can help to quickly bring people up to speed on a concept — like genetic engineering — without adding on lots of baggage.
But whenever a cell is ‘programmed,’ it’s important to consider all the crap that happens when a piece of DNA gets tweaked. Cells can behave in very strange and unpredictable ways, even when only a few changes are made to their genomes. Let me explain by giving an example that blew my mind last week: codons.
Every living thing has a genome made from DNA. Big proteins slide along these genomes at terrifying speeds, making RNA from DNA at a rate of 4,000 letters per minute. The new RNA floats in the cell until another big protein, the ribosome, latches onto it. The RNA slides through the ribosome and a brand-new protein gets built. It’s a bit like magic.
The ribosome knows which amino acids to use to build the new protein, and the order to place them in, because every three letters in the messenger RNA encodes a specific building block. ‘GAU’ codes for aspartate, ‘AAA’ for lysine. These triplets are called codons. There are 64 ways to arrange the four letters in RNA — C, U, A, G — into triplets, but there are only 20 amino acids. Thus, there is some redundancy in the genetic code; different triplets can code for the same amino acid. Okay, now you’re up to speed.
One of the beautiful things about the 21st century, this era of biology, is that we are learning just how transportable DNA really is. A gene can be taken from humans and inserted into bacteria, or vice versa. Trees can be imbued with parasitic DNA and insects with genes from algae. But every organism on Earth also has its own “codon usage table,” or triplet codes that it likes to use for each amino acid.
E. coli, a bacterium, prefers to use UUA to code for leucine. Human cells opt instead for CUG. So when a scientist takes a gene from a human and inserts it into E. coli, it’s normal to “refactor” the code so that the codons work well in the new host. This usually works well — switching a gene to ‘preferred’ codons increases the likelihood that the alien gene will produce proteins in the new host.
This process, called codon optimization, is so common amongst biologists that there are now dozens of websites offering to do it for free. Throughout all the years that I spent working at the bench, engineering living cells, I never thought twice about tweaking the codons in a gene.
But maybe I should have paid more attention. Replacing one codon for another that encodes the same amino acid — so-called synonymous mutations — can have massive effects on a protein’s function. I should have seen the warning signs. Scientists have been telling people about the dangers of codon optimization for decades, usually by penning reviews or opinion pieces in journals. But I never got the message.
There’s a rich history in virology of scientists using codon de-optimization to make viruses less able to infect and reproduce inside of cells (including for SARS-CoV-2). But attempts to improve codons can also make things worse.
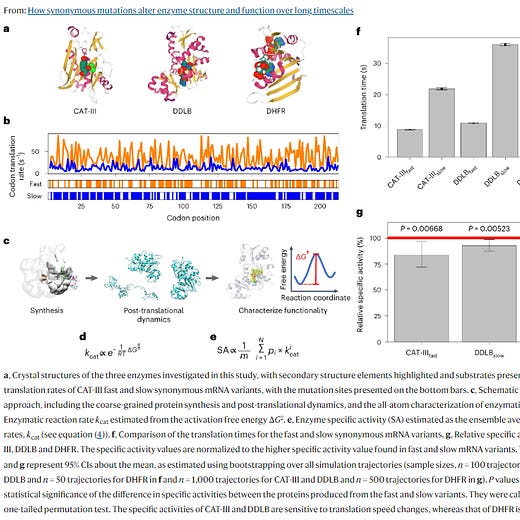
For a paper in Nature Chemistry, published earlier this month, researchers took three proteins in E. coli and, for each of them, made a fast- and slow-translated version. In other words, they tweaked the genes’ codons, using synonymous mutations, to make them zip through a ribosome faster or slower.
A fast-translated version of one enzyme had a 20 percent decrease in activity compared to its normal version. For another enzyme, the slow-translated version had about a 9 percent drop in activity. Just remember that these are the exact same protein sequences; they’re just encoded by different codons.
But it seems to be worse than just a momentary “dropoff” in activity. These effects on an enzyme can last for a really long time, like forty minutes — which is two cell divisions for some quickly-dividing microbes!
Synonymous mutations can also cause a whole host of other problems (from an excellent review article):
Although synonymous mutations were previously thought to be silent, a large body of evidence has demonstrated that codon usage can play major roles in determining gene expression levels and protein structures. Codon usage influences translation elongation speed and regulates translation efficiency and accuracy.
Changing the codons that encode a protein can also cause mRNA to decay faster or slower, and can make proteins fold faster or slower. It’s a mess. And the knock-on effects from editing codons don’t really seem predictable ahead of time (at least, not yet). Sometimes messing with codons is a big deal, and sometimes it isn’t. In other words, we don’t always understand what is happening inside a cell when we mess with its genes.
This whole fiasco reminds me of that WIRED YouTube series where an expert explains one concept in five levels of difficulty. Level one, in our codon example, is probably just “DNA makes RNA makes protein.” Level three might be “DNA makes RNA, but not always, and the RNA also breaks down in a cell, but a certain form of RNA, called messenger RNA, encodes proteins.” Level five is this codon stuff; its implications are still emerging.
But isn’t this the beauty of biology? It’s as complex as we want it to be. It can be talked about at a hundred different levels, from a thousand different angles. And as soon as we think we’ve got a basic grasp of its outlines — a vague sense of its potential — it surprises again! A wrench is thrown into the spanners!
We may never understand life in all its details, but a chaotic nature is good. Who wants all clean lines and predictable code? Here’s to biology in its infinite weirdness. ◼️
The List 🔻
Notable science from the last week.
(↑ = recommended)
AI + Compute
Accuracy and data efficiency in deep learning models of protein expression. Nikolados E-M et al. Nature Communications. Link
AI can predict how much protein will be made from a given DNA sequence with scary good accuracy, and without training on nearly as much data as previously thought.
Deep embedding and alignment of protein sequences. Llinares-López F et al. Nature Methods. Link
A language model can spot a protein homolog — a similar protein in a different organism — with three-fold higher accuracy than existing algorithms.
Deep learning models for predicting RNA degradation via dual crowdsourcing. Wayment-Steele HK et al. Nature Machine Intelligence. Link
From a crowdsourced competition emerged a model that can predict how quickly a given RNA sequence degrades within a cell. About 40% of “predictions from the winning model were within experimental error of the ground truth measurement.”
Simple nearest-neighbour analysis meets the accuracy of compound potency predictions using complex machine learning models. Janela T & Bajorath J. Nature Machine Intelligence. Link
Basic Research
A secreted effector with a dual role as a toxin and as a transcriptional factor. Wang D et al. Nature Communications. Link
There’s a specific species of bacteria that secretes a protein that controls gene expression in neighboring cells of the same species, but acts as a toxin to competitors. Very strange.
Acceleration of genome replication uncovered by single-cell nascent DNA sequencing. van den Berg J et al. bioRxiv. Link
A clever DNA sequencing method is used to measure the speed of DNA replication, or how fast a genome is copied. DNA replication speeds up up in S-phase of cell division.
Biomanufacturing
Biosynthesis of mushroom-derived type II ganoderic acids by engineered yeast. Yuan W et al. Nature Communications. Link
Baker’s yeast were genetically engineered to produce ganoderic acids, a class of chemicals produced by medicinal mushrooms. Yummy.
High-yield production of FK228 and new derivatives in a Burkholderia chassis. Gong K et al. Metabolic Engineering. Link
A drug called FK228, approved to treat T-cell lymphomas, is produced by engineered bacteria.
Engineering Pseudomonas putida KT2440 for chain length tailored free fatty acid and oleochemical production. Valencia LE et al. Communications Biology. Link
Brain
Tagging active neurons by soma-targeted Cal-Light. Hyun JH et al. Nature Communications. Link
Engineered proteins detect calcium flux in the soma of neurons and trigger gene expression in response. They can be used to record specific, active neurons in a brain.
Editing DNA
Compact Cas9d and HEARO enzymes for genome editing discovered from uncultivated microbes. Goltsman DSA et al. Nature Communications. Link
Base editors are proteins that can swap DNA letters, like ‘C’ to ‘T’. This paper reports the smallest one yet; just 632 amino acids, discovered by combing through billions of protein sequences.
CRISPR-free, programmable RNA pseudouridylation to suppress premature termination codons. Song J et al. Molecular Cell. Link
TREX1 restricts CRISPR-Cas9 genome editing in human cells. Karasu ME et al. bioRxiv. Link
This is an important paper. The higher the level of a protein called TREX1 inside a cell, the lower the CRISPR gene-editing efficiency in that cell. Removing TREX1 can improve gene editing in some human cell lines.
Engineered Cells, New Functions
Programming multicellular assembly with synthetic cell adhesion molecules. Stevens AJ et al. Nature. Link
Synthetic proteins act like LEGO pieces that can bind cells together. Arranging these proteins in specific ways was used to build three-dimensional patterns and shapes with living cells.
Targeted bacterial conjugation mediated by synthetic cell-to-cell adhesions. Robledo M et al. Nucleic Acids Research. Link
De novo engineering of a bacterial lifestyle program. Kong W et al. Nature Chemical Biology. Link
A genetic circuit is used to switch bacterial cells between a free-living and biofilm state, back and forth.
Directed evolution of generalist biosensors for single ring aromatics. Cole HO et al. bioRxiv. Link
Engineering a “detect and destroy” skin probiotic to combat methicillin-resistant Staphylococcus aureus. Guan C et al. PLOS One. Link
Health
A single-shot ChAd3-MARV vaccine confers rapid and durable protection against Marburg virus in nonhuman primates. Hunegnaw R et al. Science Translational Medicine. Link
Marburg virus kills about 90 percent of the people it infects. There have been a few recent cases in West Africa. This phase I trial of a vaccine, tested in chimpanzees, shows protective immunity for at least one year.
Optimization of the proliferation and persistency of CAR T cells derived from human induced pluripotent stem cells. Ueda T et al. Nature Biomedical Engineering. Link
Synthetic cytokine circuits that drive T cells into immune-excluded tumors. Allen GM et al. Science. Link
Trial of Beremagene Geperpavec (B-VEC) for dystrophic epidermolysis bullosa. Guide SV et al. The New England Journal of Medicine. Link
Phase III trial with 31 people tested whether a skin cream can help treat wounds caused by a rare, genetic disease. Herpes simplex viruses, in the cream, deliver a collagen gene, called COL7A1, into the skin. About two-thirds of wounds treated with the gene therapy were completely healed at 6 months, compared to 22 percent of those treated with placebo.
A universal influenza mRNA vaccine candidate boosts T cell responses and reduces zoonotic influenza virus disease in ferrets. Van de Ven K et al. Science Advances. Link
Hearables: Feasibility of recording cardiac rhythms from single ear locations. Yarici M et al. bioRxiv. Link
An accurate ECG reading of the heart was taken by placing sensors near the ear. This paper opens the door for health-monitoring earbuds. Like an Apple Watch but for your ear.
Suppression of flavivirus transmission from animal hosts to mosquitoes with a mosquito-delivered vaccine. Wen D et al. Nature Communications. Link
Sequence-specific capture and concentration of viral RNA by type III CRISPR system enhances diagnostic. Nemudraia A et al. Nature Communications. Link
A sensitive diagnostic test for SARS-CoV-2 uses CRISPR gene editing to detect the virus on nose swabs, with a sensitivity down to just 15 fM.
Gene augmentation prevents retinal degeneration in a CRISPR/Cas9-based mouse model of PRPF31 retinitis pigmentosa. Xi Z et al. Nature Communications. Link
Yet another gene therapy to prevent retinal degradation caused by retinitis pigmentosa. This one, in a mouse model, preserved vision after mote than 10 weeks.
An implantable soft robotic ventilator augments inspiration in a pig model of respiratory insufficiency. Hu L et al. Nature Biomedical Engineering. Link
The potency and synergy of plant-made monoclonal antibodies against the BA.5 variant of SARS-CoV-2. Sun H et al. Plant Biotechnology Journal. Link
Tobacco plants were genetically engineered to make monoclonal antibodies that can partially neutralize a few different Omicron variants.
CRISPR-based diagnostics: Challenges and potential solutions toward point-of-care applications. Ghouneimy A et al. ACS Synthetic Biology. Link
Protein & Molecular Engineering
Designer installation of a substrate recruitment domain to tailor enzyme specificity. Park R et al. Nature Chemical Biology. Link
A protein was designed, on a computer, to interact with other proteins and modulate their activities. It’s sort of like an ‘enhancer’ to tune an enzyme’s function.
Turning antibodies off and on again using a covalently tethered blocking peptide. Brasino M et al. Communications Biology. Link
Another one: A de novo designed protein binds to antibodies and prevents them from attaching to a target. But when these proteins are struck by light, they fall off and the antibody switches ‘on’.
A de novo protein catalyzes the synthesis of semiconductor quantum dots. Spangler LC et al. PNAS. Link
A genetically encoded fluorescent sensor for manganese(II), engineered from lanmodulin. Park J et al. PNAS. Link
Science and Society
Nomenclature of cell-cultivated meat & seafood products. Malerich M & Bryant C. npj Science of Food. Link
Naming foods makes a big difference in terms of whether or not they’re accepted by shoppers. The word “artificial” is particularly bad, and make people think a food is less safe than something that was “cultivated.”
Consumer acceptance of new plant-breeding technologies: An application to the use of gene editing in fresh table grapes. Uddin A et al. PLOS One. Link
Making a Cell
L-SCRaMbLE creates large-scale genome rearrangements in synthetic Sc2.0 chromosomes. Lindeboom TA. bioRxiv. Link
Yeast cells carrying a synthetic genome had their DNA ‘scrambled,’ or shuffled around, when exposed to light. DNA sequencing was then used to find genome variants with new functions.
Dynamic assembly of DNA-ceria nanocomplex in living cells generates artificial peroxisome. Yao C et al. Nature Communications. Link
The Wild
Parallel engineering of environmental bacteria and performance over years under jungle-simulated conditions. Chemla Y et al. PLOS One. Link
Engineered bacteria, grown in conditions that replicate a jungle, were viable and still performed their new functions after 21 months of storage!
Tools & Technology
Recent advances in crop transformation technologies. Chen Z et al. Nature Plants. Link
This review highlights the many improvements we’ve recently made to plant engineering. It has never been easier to make crops with bigger roots or higher yields.
PEAC-seq adopts Prime Editor to detect CRISPR off-target and DNA translocation. Yu Z et al. Nature Communications. Link
Achieving single nucleotide sensitivity in direct hybridization genome imaging. Wang Y et al. Nature Communications. Link
Other Cool Stuff
Gigapixel imaging with a novel multi-camera array microscope. Thomson E et al. eLife. Link
A gigapixel microscope made from 96 smartphone cameras. Computer code stitches together the recordings and the cameras have a resolution down to 18 microns.
How to grow (almost) anything: a hybrid distance learning model for global laboratory-based synthetic biology education. Perry E et al. Nature Biotechnology. Link
MIT has a remote course where students around the world learn how to engineer biology. Last year, students made a “portable bioreactor for do-it-yourself biosensor testing" and "engineered bacteria to produce biological art, remotely...patterned via a lab robot in the U.S. by a student in Germany." Genetic engineering goes global!
Musings on the Future
Tweets, essays & news articles about the future.
The craziest thing I watched last week. ⛛ What if biologists could design proteins or entire experiments, together, using projections cast upon a table? Clever re-imagining of the laboratory by Bret Victor and Dynamicland. (h/t @stevekrouse)
Forget the new Avatar movie. “Make People Better” comes out tomorrow on Prime Video and, apparently, has lots of new footage and audio recordings from the CRISPR Baby Fiasco. The movie “casts doubt on the dominant narrative of Chinese scientist He Jiankui” — who was recently released from prison — “as a ‘rogue’ actor.” STAT
Massive news from Moderna. An mRNA-based cancer vaccine, given in combination with Keytruda, an antibody used to treat certain cancers, reduced risk of melanoma recurrence or death by 44 percent compared to Keytruda alone in a phase II clinical trial. Press release
Noah Smith’s “techno-optimist” musings on what might come in 2023; most of it is energy and AI. Noahpinion
“Why the age of American progress ended,” by Derek Thompson, argues that America is good at inventing stuff, but not scaling it. The argument seems to apply to biotechnology, too, at least in terms of academic-to-company spinouts. How many research papers are published each year, with promising real-world applications, but are never taken forward? There are holes in the VC market, especially for consumer biotechnology. The Atlantic



Great essay as usual and very useful info. Real science as opposed to the low quality junk pop sci proliferating today. Thanks Codon.