You’re reading Codon, a newsletter about biology, technology & the future. Thank you for being here. ❤️
A bacterium’s genome, pulled into a straight thread, is nearly 1,000 times longer than the cell from which it came. If you placed one E. coli into a plastic, gallon carton and waited a few hours, the genomes of its descendants, placed end-to-end, would reach to the moon and back.1
One rarely pauses to ponder how so much DNA — let alone sugar, proteins, and everything else — can fit inside such a small vessel. Biochemistry textbooks depict cells as spacious places, where molecules float in secluded harmony. “But a cell looks more like a burrito,” says Michael Elowitz, a biologist at Caltech. All the biochemicals are pushed together and bumping into each other. It’s a wonder, then, that anything ever gets done inside of living cells, for they are very fast and crowded places. A David Goodsell painting enamors because it conveys this denseness in visual form.

Photographs and paintings are beautiful, but ultimately simplistic. They are snapshots in time. Pictures can hint at complexity, but don’t convey the intense dynamics of a living cell. All of our scientific methods to study life, similarly, demand that cells be killed or frozen before a microscopic image is taken. Mathematics and words are our best tools to consider the active chaos of living cells.
For years, though, I had an intense aversion to mathematics. Biology was my refuge because it was simple: Read the textbook, memorize the facts, ace the exam. (I majored in biochemistry because it didn’t have a multivariable calculus requirement). And then I started a PhD at Caltech and landed in Rob Phillips’ lab. Suddenly, I was thrown into the deep end of biophysics. I took courses like Physical Biology of the Cell and wrote out statistical mechanics equations on big whiteboards. I felt like a scientist for the first time, and I began to grasp the “numbers of biology” (many calculations in this essay come from that course and another textbook, Cell Biology by the Numbers.)
Without mathematics, I learned, biology is naked; we can only comprehend it at arm's length. But with numbers, living cells come alive. Mathematics enables us to see a David Goodsell painting with fresh eyes.
Just consider the Central Dogma. Students learn the basics through words: DNA is transcribed to RNA, which is translated into proteins. But what does this really mean? How fast does DNA become RNA, or RNA protein? How many proteins are in a cell, and how fast do they actually move? Doing these calculations reveals both the beauty and weirdness of life at the smallest scales. It lends a deeper appreciation to biology. And all we need is a pencil and paper.
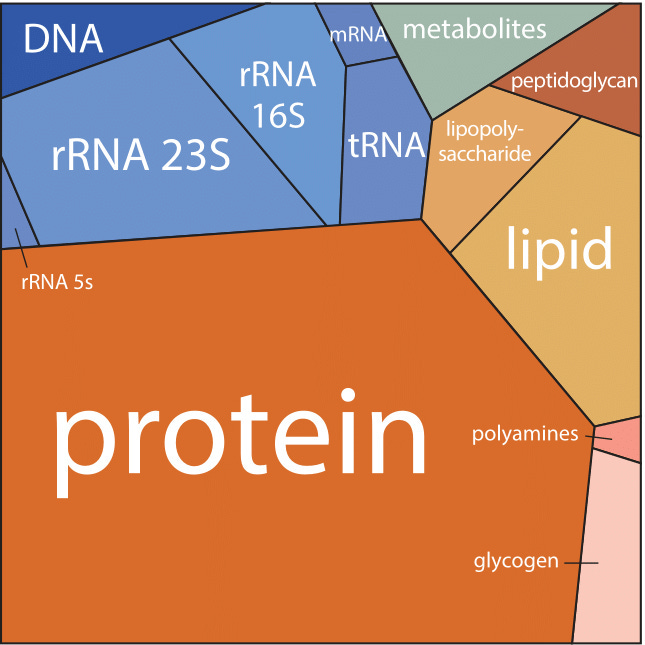
But first, some background. A microbe’s guts are a veritable Times Square, crowded with sugars, proteins, and water molecules that ricochet and smash into each other billions of times each second. Space is limited. A bacterium’s insides are 70 percent water by mass. The other 30 percent is dominated by proteins first, followed by RNA and lipids. DNA accounts for just one percent. All of this stuff fits inside a volume that is one-quadrillionth of a liter. (500 billion microbes fit inside of an Aspirin tablet.)
Let’s first think about DNA to RNA, or transcription. A typical E. coli has 4,400 genes and, at any given moment, about 25 percent of them are being copied into RNA by a large protein, called RNA polymerase. These proteins latch onto a gene and zip along at breakneck speeds, converting 40 bases every second. If this enzyme was scaled up to the size of a human, its speed would be double that of Usain Bolt’s pace in a 100-meter dash.2 The protein makes about one mistake for every 100,000 letters.
Less than 30 seconds pass between the time that the protein binds to a DNA sequence and finishes making an RNA. And as soon as the RNA is complete, a small army of ribosomes swoop in and latch onto it. These proteins read the letters in the RNA sequence — three at a time — and convert them into amino acids in a growing protein.
Ribosomes also move very quickly. They can make a typical protein from RNA in just 24 seconds. A single ribosome could translate the first Harry Potter book in two-and-a-half hours while making about three dozen typos along the way.3
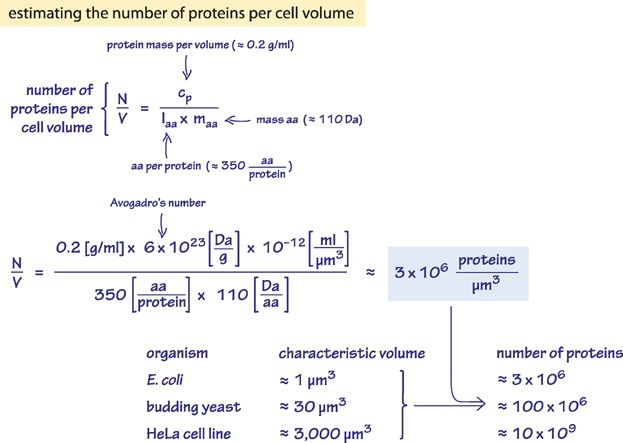
When a ribosome has finished its job, its jaws unclench, the new protein is released, and it quickly diffuses away. A typical cell contains three or four million proteins that break down sugar, copy DNA, or send signals to nearby cells. Biology is an autonomous factory, where machines build machines that build themselves.
At the small scales in which proteins operate, strange things also begin to happen. Diffusion is a prime example.
Very small molecules, like water or ions, diffuse very fast; up to one centimeter each second. But large proteins move more slowly. They float along, from one place to the next, and travel just a few micrometers (one-millionth of a meter) in the same time span. Diffusion is given in units of length2/time, which means that it would take a protein 10 milliseconds to move across a cell, but 20 days to travel one centimeter.
Diffusion sets an upper limit on a cell’s size.4 If a cell is too small, not enough ‘stuff’ fits inside and evolution is constrained. If a cell is too large, nothing ever gets done because proteins never reach their destination. Life, then, is a search for the Goldilocks principle.
As proteins move through a cell, they are also bombarded by water, sugars, and other proteins. In biology textbooks, one often hears sentences like, “A protein’s substrate has a concentration of 0.5 millimolar.” This means that, inside the cell, there is one substrate for every 100,000 water molecules, and the enzyme collides with 500,000 substrates every second. Proteins lock onto their target — and ignore everything else — by forming many weak, noncovalent bonds to a substrate in a fraction of a second.
Cells, then, are basically just chaotic swarms of energy and fortuitous accidents. The Central Dogma sounds simple in words, but is a miracle in reality. It’s a wonder that cells get anything done at all.
The first time I did these calculations, I felt an intense appreciation for biology. And now, I want young students to feel the same. We need to teach biologists to think as mathematicians; to carefully quantify biology, to think in absolute units, and to develop a feeling for the organism.
Another thing: Throughout this essay, I’ve depicted cells as dense blobs filled with lots of stuff. This insinuates that, if one studied everything in the cell and tallied all of its components, then perhaps we’d have a complete knowledge of biology. But this isn’t true.
Some proteins, for instance, “moonlight” in the cell. They carry out one function when their substrate is around, and do something entirely different when it isn’t. Many protein signaling pathways also play a specific role in one type of cell, and something different in another, as Michael Elowitz’s group at Caltech has recently shown. Biology is infinitely weird. If we ever plan to master it, we will need new scientific methods to measure protein dynamics and interaction strengths.
When COVID came in 2020, I left my Ph.D. and moved to New York to study journalism. I fell out of contact with Rob, but my appreciation for biological numbers remained. I still enjoy jotting down little calculations on the back of an essay draft. And every day, I still feel grateful that I get to learn about biology, a field that is far stranger than anything one could see while scuba diving or traveling to Mars.
It is still difficult for me to imagine the micro, when I only experience the macro. But a pen, paper, and imagination seem to suffice.
Thanks to Tony Kulesa, Yonatan Chemla & Avadhoot Jadhav for feedback.
The views expressed in this blog are entirely my own and do not represent the views of any company with which I am affiliated. All mistakes are my own.
Correction: The calculation in the first paragraph was off by a factor of 10. The E. coli genome stretches about 0.17 centimeters in length, with a distance of about 0.34 nanometers between bases in DNA. Thank you to David Savage for the correction. The burrito analogy was initially coined by Michael Elowitz, about two decades ago.
Appendix
An E. coli genome contains about 5 million bases of DNA. Every letter, in that genome, is 0.34 nanometers away from adjacent letters. This means that the physical length of a straightened genome is 0.17 centimeters, or roughly 1,000 times longer than the length of an E. coli cell (about 2 microns).
The moon is 384,400 kilometers away from Earth and so it would take 2.26 x 1011 E. coli genomes, placed end to end, to reach it and twice that number to return to Earth. A saturated growth media contains one billion cells per milliliter.RNA polymerase is physically about 10 nanometers wide. Each base in DNA is 3.4 nanometers apart, and so RNA polymerase sits atop roughly 3 nucleotides at once. If RNA polymerase transcribes 40 nucleotides per second, then it is moving 136 nanometers in that timespan, or 13.5 times more than its ‘stride length.’ A typical human stride length is 2.3 feet while walking, or about 5 feet while running (h/t Danielle Steffen). If we multiply by 13.5 a pace that is roughly double the top speed of Usain Bolt’s world record sprint.
There are 77,000 words in the first Harry Potter book. The average English word has 5 letters, which implies that the book has somewhere around 385,000 letters. A ribosome reads 42 RNA nucleotides per second, which means it could read all letters in the book in about 152 minutes. The ribosome’s error rate is about 1 in 10,000, but this varies quite a bit based on the sequence.
Of course, there are exceptions to every rule (especially in biology). The giant squid axon has a diameter of 1 millimeter and is nearly a meter in length!



Hi. Niko. This is a wonderful writeup. Can I get permission to translate it to Kannada, a language of 80 million Indians? I edit a freely distributed monthly where I would like to publish this. My podcast science digest is here https://youtu.be/0cSYsV0PbBs?feature=shared
As an aspiring PhD student and predominantly based in Biology I have discovered a new found passion for mathematics and its usage in Biology. Thanks for re-inforcing this belief Niko. Loved this....!!