A Light Switch for Gene Editing - 2021.03.08
Plus: All the other research in synthetic biology this week.
☀️ Good morning.
Seen in the light of evolution, biology is, perhaps, intellectually the most satisfying and inspiring science. Without that light it becomes a pile of sundry facts — some of them interesting or curious but making no meaningful picture as a whole.
—Theodosius Dobzhansky
A Light Switch for Gene Editing: I’m a sucker for CRISPR, but it has its drawbacks. The Cas9 protein chomps away at DNA until it is removed, or its guide RNA is destroyed. CRISPR-Cas9 can also exert a “burden” on cells (perhaps due to off-target cutting of DNA), causing them to grow more slowly.
There are a lot of ways to shut down Cas9, but none are ideal. Anti-CRISPR proteins can switch off DNA-editing, for example, but they have to be expressed in the cell at a Goldilocks level. Chemicals can also be used, but that demands careful dosage to avoid harming the cells.
For a new study in Molecular Cell, titled “Cas9 deactivation with photocleavable guide RNAs,” researchers at Johns Hopkins University created a new technology to shut down Cas9.
Their approach is simple: Within the crRNA portion of a guide RNA—the bit that actually binds to the DNA target—they implanted an internal, photocleavable linker molecule, called 2-nitrobenzyl. The linker molecule is placed five bases away from the end of the crRNA, and if you blast the cells with light (365 nm wavelength), the linker is obliterated and the end of the guide RNA falls off. Cas9 stops cutting.
This photocleavable approach is very fast, with nearly total Cas9 deactivation within one minute of light exposure. The authors write that their approach is “the fastest and most complete strategy for Cas9 deactivation, improving on prior arts by at least an order of magnitude in both speed and residual activity.”
Why It Matters: Methods to switch off Cas9 have historically worked by adding an additional component to cells—anti-CRISPR proteins, chemicals, siRNA sequences, and so forth. This new approach for Cas9 deactivation is non-invasive, requiring only light and a modified guide RNA. It could be used to reduce the toxicity that Cas9 exerts on living cells, or to answer gene-editing questions that have a temporal or spatial component.
This study does have an obvious limitation, though. Since the guide RNAs are chemically modified to carry a photocleavable linker, they cannot be genetically-encoded in the cells, as far as I know. They must be electroporated into the cells.
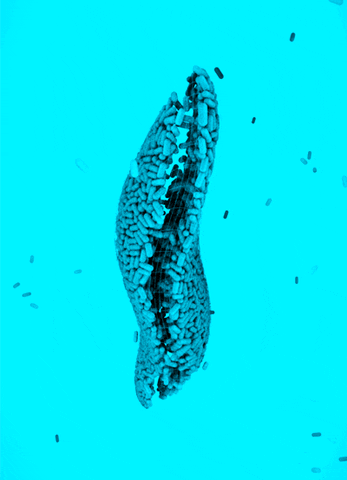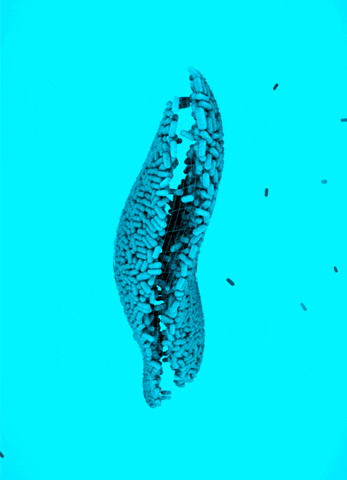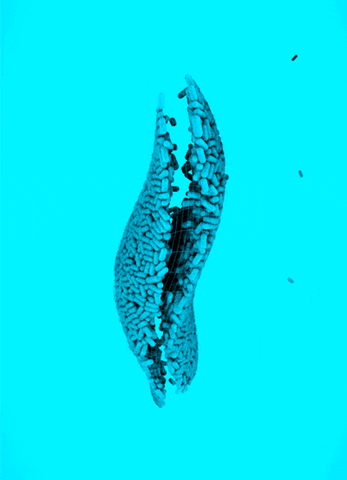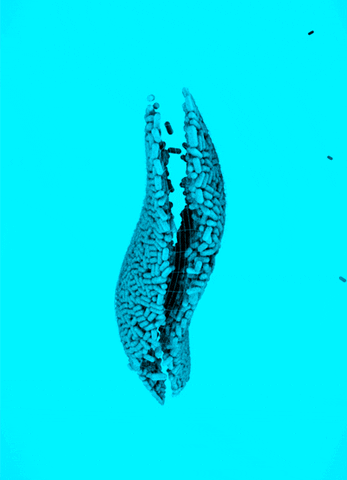
CRISPR Chickens, CRISPY Pigs: For a new study in PNAS, titled “Cas9-expressing chickens and pigs as resources for genome editing in livestock,” researchers at the Technical University Munich created pigs and chickens that endogenously express Cas9 in all of their organs. The Cas9-packing animals were “healthy and fertile.”
Cas9 transgenic pigs were created by inserting the Cas9 gene—from Streptococcus pyogenes—into a specific, genetic locus in pig cell clones, called ROSA26. The researchers then performed a somatic cell nuclear transfer, leading to the live birth of two piglets. Chickens carrying Cas9, also from S. pyogenes, were made in a different way, but with similar results.
To demonstrate the utility of these Cas9-carrying animals, the researchers targeted many different genes in various cell types, including lymphocytes, the developing brain, and the heart. They tested several approaches for delivering the guide RNAs, including “transfection with synthetic gRNAs, in ovo electroporation, or viral-based delivery methods;” all of them worked, albeit with varying efficiencies.
Why It Matters: Pigs and chickens have been genetically-engineered with CRISPR-Cas9 for years. This study lowers the barrier of entry for editing these agriculturally-relevant animals. Researchers that use these animals will not need to deliver both Cas9 and guide RNAs to edit a gene; they only have to deliver a guide RNA, which can be easily packaged inside of viral vectors. This work could also speed up the design and creation of livestock animals that have improved features, an ethically precarious effort that, despite some objections, is already underway.
Plasmids Are So 2020: If you take some cells, grow them in an incubator, and measure their gene expression, what are you actually measuring? Answer: the mean expression for millions (or billions) of cells; bulk measurements obscure the individual variability that exists between them.
Synthetic biologists routinely make bulk measurements, but a new study from Chris Voigt’s group, at MIT, suggests that they should do so with caution. The MIT team developed a method that can “simultaneously count plasmid DNA, RNA transcripts, and protein expression in single living bacteria” by using fluorescent reporters, fused to binding proteins, to label plasmids and RNA transcripts. This approach enables both plasmids and RNA transcripts to be counted inside of single cells. The new paper is titled “Single-cell measurement of plasmid copy number and promoter activity,” and was published in Nature Communications
Using their method, the authors found “shockingly high cell-to-cell variability” in how many copies of a plasmid bacteria actually receive. They also found that, when transfecting plasmids into cells, a large percentage of cells don’t receive any plasmid at all, even when they selected for antibiotic resistance. Voigt tweeted about the results on March 5th, and called for researchers to “stop using” plasmids, presumably in certain contexts.
Why It Matters: A mechanical engineer would never build a bridge without first taking careful measurements. While building a cell is not as easy as building a bridge (yes, I really wrote that), synthetic biologists should still strive to take the same precision measurements as other engineering disciplines. This new method can help researchers directly observe cell-to-cell variations, possibly improving those measurements.

This thread, from the Lopatkin lab, is worth your time. For a new study, in Molecular Systems Biology, her group found that some plasmids are more difficult for bacteria to acquire than others. 👇
🧫 Other Studies Published This Week
Am I missing coverage on a certain topic? Please leave a comment on this post.
Artificial Life
Experimental tests of functional molecular regeneration via a standard framework for coordinating synthetic cell building. bioRxiv. Preprint. Link
Emergence of function from single RNA sequences by Darwinian evolution. bioRxiv. Preprint. Link
Biomaterials
Bacterial cellulose retains robustness but its synthesis declines after exposure to a Mars-like environment simulated outside the International Space Station. Astrobiology. Link
Biosensors
A multiplexed, automated evolution pipeline enables scalable discovery and characterization of biosensors. Nature Communications. Open Access. Link
Guanidine biosensors enable comparison of cellular turn-on kinetics of riboswitch-based biosensor and reporter. ACS Synthetic Biology. Open Access. Link
DNA Storage & Nanotechnology
DNA stability: a central design consideration for DNA data storage systems (Review). Nature Communications. Open Access. Link
Fundamental Discoveries
Systematic discovery of pseudomonad genetic factors involved in sensitivity to tailocins. The ISME Journal. Open Access. Link
Conjugation dynamics depend on both the plasmid acquisition cost and the fitness cost. Molecular Systems Biology. Open Access. Link
Streamlining CRISPR spacer-based bacterial host predictions to decipher the viral dark matter. Nucleic Acids Research. Open Access. Link
Systematic analysis of factors that improve homologous direct repair (HDR) efficiency in CRISPR/Cas9 technique. PLOS ONE. Open Access. Link
Gene Drives
Inherently confinable split-drive systems in Drosophila. Nature Communications. Open Access. Link
Genetic Engineering & Control
Programmable C:G to G:C genome editing with CRISPR-Cas9-directed base excision repair proteins. Nature Communications. Open Access. Link
Identification of permissive amber suppression sites for efficient non-canonical amino acid incorporation in mammalian cells. Nucleic Acids Research. Open Access. Link
Random and combinatorial mutagenesis for improved total production of secretory target protein in Escherichia coli. Scientific Reports. Open Access. Link
Medicine and Diagnostics
Quantifying the limits of CAR T-cell delivery in mice and men. Journal of the Royal Society Interface. Open Access. Link
An in vivo method for diversifying the functions of therapeutic antibodies. PNAS. Link
Point-of-care bulk testing for SARS-CoV-2 by combining hybridization capture with improved colorimetric LAMP. Nature Communications. Open Access. Link
A multiplexed, next generation sequencing platform for high-throughput detection of SARS-CoV-2. Nature Communications. Open Access. Link
Targeted-antibacterial-plasmids (TAPs) combining conjugation and CRISPR/Cas systems achieve strain-specific antibacterial activity. Nucleic Acids Research. Open Access. Link
Metabolic Engineering
Reversible thermal regulation for bifunctional dynamic control of gene expression in Escherichia coli. Nature Communications. Open Access. Link
DIVERSIFY: A fungal multispecies gene expression platform. ACS Synthetic Biology. Link
Peroxisomes: a new hub for metabolic engineering in yeast (Opinion). Frontiers in Bioengineering and Biotechnology. Open Access. Link
Pathway discovery and engineering for cleavage of a β-1 lignin-derived biaryl compound. Metabolic Engineering. Link
Engineered polyploid yeast strains enable efficient xylose utilization and ethanol production in corn hydrolysates. Frontiers in Bioengineering and Biotechnology. Link
Engineering heterologous molybdenum-cofactor-biosynthesis and nitrate-assimilation pathways enables nitrate utilization by Saccharomyces cerevisiae. Metabolic Engineering. Open Access. Link
Partitioning metabolism between growth and product synthesis for coordinated production of wax esters in Acinetobacter baylyi ADP1. Biotechnology & Bioengineering. Link
New Technology
Engineered yeast genomes accurately assembled from pure and mixed samples. Nature Communications. Open Access. Link
Chemogenetic ON and OFF switches for RNA virus replication. Nature Communications. Open Access. Link
Dual DNA and protein tagging of open chromatin unveils dynamics of epigenomic landscapes in leukemia. Nature Methods. Link
Luciferase-based reporter system for in vitro evaluation of elongation rate and processivity of ribosomes. Nucleic Acids Research. Open Access. Link
Microbial Communities
Directed evolution of microbial communities. Annual Review of Biophysics. Open Access. Link
Optogenetics
Using a bistable animal opsin for switchable and scalable optogenetic inhibition of neurons. EMBO Reports. Open Access. Link
Optogenetic modification of Pseudomonas aeruginosa enables controllable twitching motility and host infection. ACS Synthetic Biology. Link
Plants
Developing a Nicotiana benthamiana transgenic platform for high‐value diterpene production and candidate gene evaluation. Plant Biotechnology Journal. Open Access. Link
Seeing is believing: a visualization toolbox to enhance selection efficiency in maize genome editing. Plant Biotechnology Journal. Open Access. Link
Advanced domestication: harnessing the precision of gene editing in crop breeding (Review). Plant Biotechnology Journal. Open Access. Link
Generation of novel self‐incompatible Brassica napus by CRISPR/Cas9. Plant Biotechnology Journal. Open Access. Link
Protein Engineering
Enzyme discovery and engineering for sustainable plastic recycling (Review). Trends in Biotechnology. Link
A universal chemical method for rational design of protein-based nanoreactors. bioRxiv. Preprint. Link
De novo design of a reversible phosphorylation-dependent switch for membrane targeting. Nature Communications. Open Access. Link
Systems Biology & Modelling
Metabolic fitness landscapes predict the evolution of antibiotic resistance. Nature Ecology and Evolution. Link
Miscellaneous Topics
Applications, challenges, and needs for employing synthetic biology beyond the lab (Perspective). Nature Communications. Open Access. Link
No small matter. (Meeting report from the 2020 Nanopore Electrochemistry Meeting.) Nature Chemistry. Open Access. Link
Until next time,
— Niko
Thanks for reading Cell Crunch, part of Bioeconomy.XYZ. If you enjoy this newsletter, please share it with a friend or colleague. A version of these newsletters is also posted on Medium. Reach me with tips and feedback on Twitter @NikoMcCarty or via email.


